Figure 1.
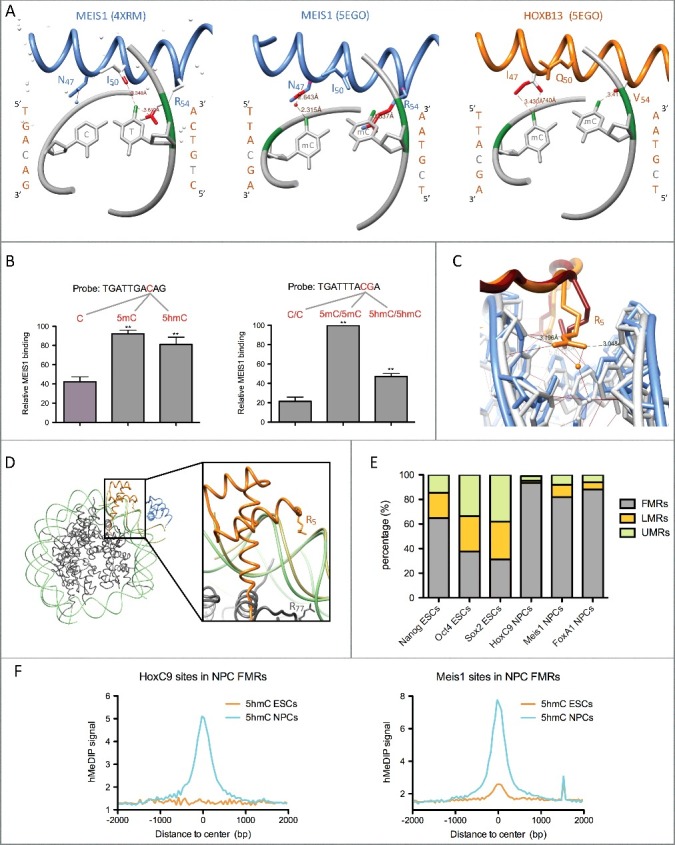
Homeodomain transcription factor interaction with methylated and hydroxymethylated DNA. (A) HD recognition helix contacts with CpA (4XRM) or 5mCpGs (5EGO). Contacts between the T or 5mC methyl groups (coloured in green) and HD residues are coloured in red. Contacts shown in the middle panel are predicted by the superimposition of the MEIS1 recognition helix to the one of HOXB13 bound to methylated DNA (5EGO), whereas the ones shown in the left and right panels are those observed in the 4XRM and 5EGO crystal structures. All structures were visualized with the UCSF Chimera 1.4.1 software. (B) In vitro pull-down assays showing binding preference of MEIS1 from NPC nuclear extracts for 5mC and 5hmC in two different DNA probes. Experimental data are from Mahé et al. [19] (C) Insertion of residue R5 within the minor groove in crystals of HOXB13 bound either to an unmethylated (5EG0, HOXB13 in red and DNA in blue) or to a methylated (5EGO, HOXB13 in orange and DNA in gray) TTACGA motif. (D) Superimposition of the crystal structures of HOXB13 (orange)/MEIS1 (blue) heterodimer bound to methylated DNA (kaki – 5EGO) and a nucleosome formed on methylated DNA (histones in dark gray and DNA in light green – 5B2J). The right panel shows an enlargement of the superimposed structures with the widened minor groove of the nucleosomal DNA likely unable to stabilize R5 insertion. (E) Distribution of TF binding sites in fully- (FMRs), low- (LMRs) and un-methylated regions (UMRs) of the mouse genome from ESCs and NPCs according to Stadler et al. [9] Mesi1 and FoxA1 binding sites are from Mahé et al. [19] NCBI GEO datasets for TF binding sites are as follows: GSM766061 (HoxC9), GSM1436068 (Nanog), GSM1436067 (Oct4), and GSM1436070 (Sox2). (F) Average ESC (GSM978374) and NPC (GSM978376) 5hmC profiles (hMeDIP-seq) centered on HoxC9 and Meis1 binding sites overlapping NPC FMRs.
