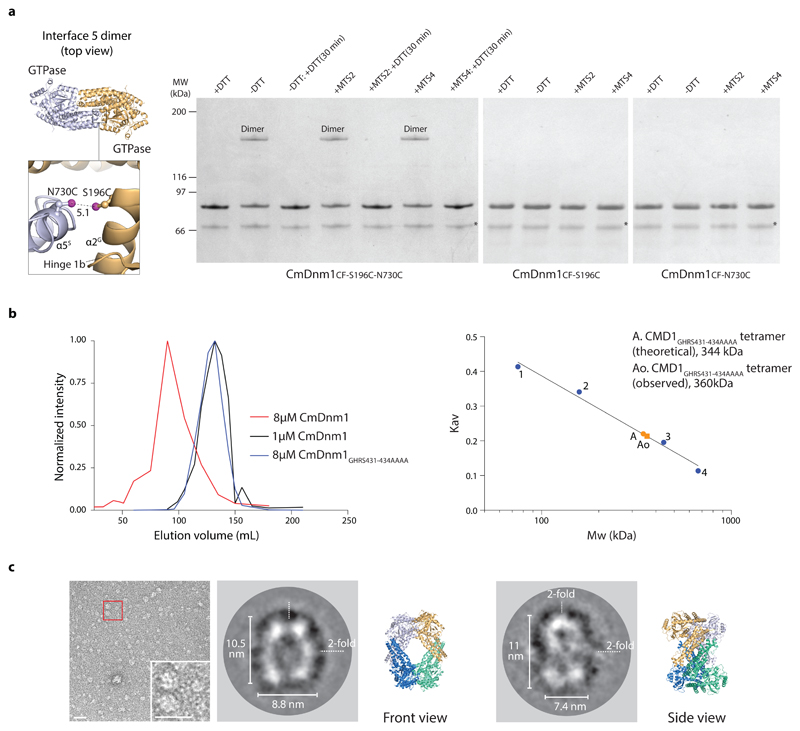
Figure 5. Validation of the CmDnm1 tetramer in solution.
a, CmDnm1CF-S196C-N730C cross-linking (S-S distance = 5.1 Å) forms a dimer in non-reducing conditions, or with MTS cross linkers. No higher order oxidation oligomers are observed. Cross-linking is not observed in the single cysteine mutants CmDnm1CF-S196C and CmDnm1CF-N730C. Cross-linking is reversible upon incubation with DTT after 30 mins reaction time. The experiment was repeated n=3 times with similar results. MW = molecular weight. b, SEC analysis of CmDnm1 oligomeric state based on concentration. At 8.0 µM, CmDnm1 is polymerised and elutes in the void volume. At 1.0 µM, CmDnm1 elutes as a tetramer (Kav = 0.25). At 8.0 µM, the self-assembly limiting stalk mutant GHRS431-434AAAA elutes as a tetramer. Calibration data includes conalbumin 75 kDa (1), aldolase 158 kDa (2), ferritin 440 kDa (3), thyroglobulin 669 kDa (4). c, Negative stain EM showing CmDnm1 single particles at low concentration (1.0 µM) (left). Class averages derived from 7740 particles typically show diamond or oval shaped particles (middle), or figure-of-8 shapes (right) that are consistent with front or side views of the CmDnm1 tetramer structure.