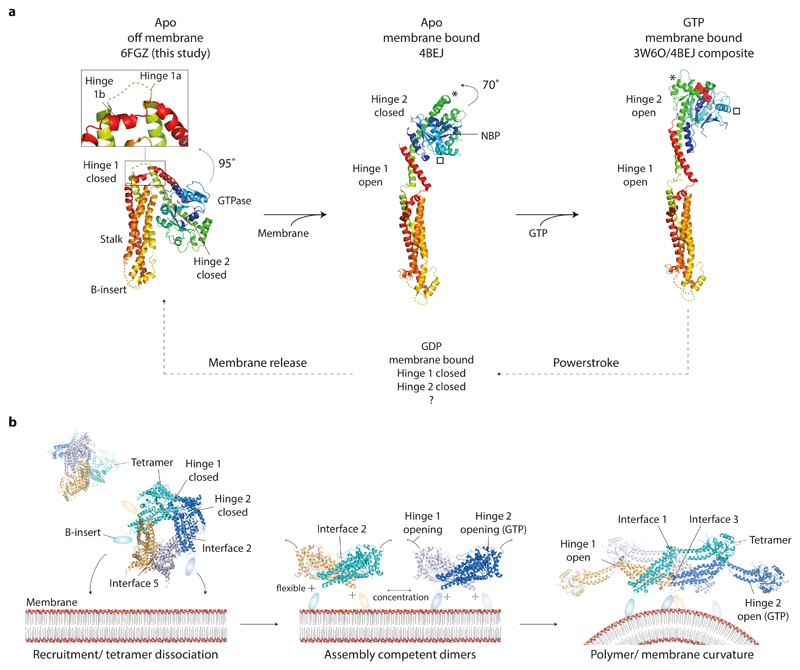
Figure 6. CmDnm1 nucleotide hydrolysis cycle and activation model.
a, General model of membrane and GTP induced conformational changes in CmDnm1 and DNM1-L. In all panels, the equivalent orientation of the stalk is shown. (Left) Whilst off membrane and in the apo state, CmDnm1 is in a hinge 1 and hinge 2 closed conformation. (Middle) Membrane binding or self-assembly induces hinge 1 opening. In the absence of nucleotide, hinge 2 remains closed as shown by the DNM1-L crystal structure (PDB 4BEJ). (Right) Composite model of DNM1-L G-domain/BSE truncation bound to GMPPCP (PDB 3W6P) with the DNM1-L stalk (PDB 4BEJ). GTP binding induces hinge 2 opening and facilitates G-dimerization. * and show the movement of equivalent structural positions between apo (middle) and GTP bound (right) states. GTP hydrolysis to GDP induces hinge 2 closure and speculatively hinge 1 closure. b, Model for CmDnm1 activation and polymerisation. (Left) CmDnm1 is off membrane and sequestered as an interface 2 and interface 5 tetramer. (Middle) Membrane recruitment yields interface 2 dimers and hinge 1 opening. (Right) Local concentration of interface 2 dimer promotes polymer formation. GTP binding is required to induce hinge 2 transition to the open conformation.