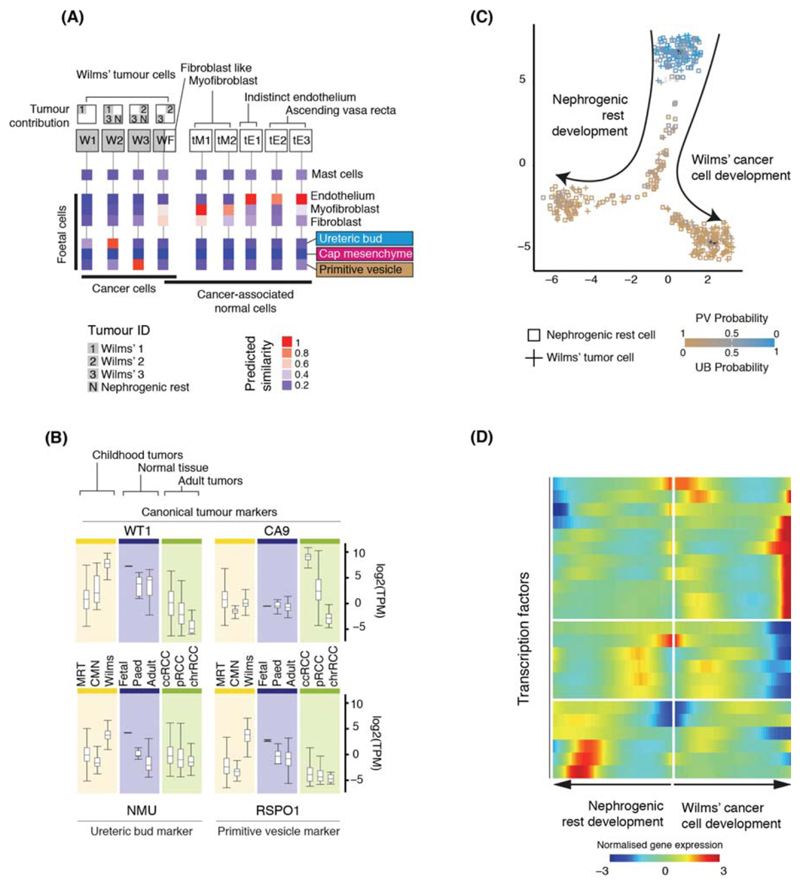
Figure 3. Matching childhood tumors with normal fetal cells.
(A) Similarity of Wilms’ tumor and cancer-associated normal cells to the reference fetal kidney map (Fig. 2A), with mast cells added as a negative control. Square boxes indicate sample contribution. Colors represent the probability that the cluster identified in the column header is “similar” to the fetal cluster identified by the row label (2).
(B) Expression of canonical tumor markers and representative UB and PV specific genes (Table S8) in RNA-seq from childhood cancers (yellow), normal tissue (blue) or adult cancers (green). MRT: malignant rhabdoid tumor; CMN: congenital mesoblastic nephroma. As positive controls, canonical tumor markers are shown: WT1 (Wilms’); CA9 (ccRCC).
(C) Pseudo-time trajectory of all Wilms tumor and nephrogenic rest cell. Color indicates similarity of each cell to the PV or UB fetal population. Jitter has been added to each point’s position with the original position plotted underneath in black (2).
(D) Transcription factors identified as varying significantly along the pseudo-time trajectory in (C). The center of the heatmap corresponds to the cells at the top of (C) and then proceeding left/right along the arrows shown in (C).