Abstract
The Adverse Outcome Pathway (AOP) framework describes the progression of a toxicity pathway from molecular perturbation to population-level outcome in a series of measurable, mechanistic responses. The controlled, computer-readable vocabulary that defines an AOP has the ability to, automatically and on a large scale, integrate AOP knowledge with publically available sources of biological high-throughput data and its derived associations. To support the discovery and development of putative (existing) and potential AOPs, we introduce the AOP-DB, an exploratory database resource that aggregates association relationships between genes and their related chemicals, diseases, pathways, species orthology information, ontologies, and gene interactions. These associations are mined from publically available annotation databases and are integrated with the AOP information centralized in the AOP-Wiki, allowing for the automatic characterization of both putative and potential AOPs in the context of multiple areas of biological information, referred to here as “biological entities”. The AOP-DB acts as a hypothesis-generation tool for the expansion of putative AOPs, as well as the characterization of potential AOPs, through the creation of association networks across these biological entities. Finally, the AOP-DB provides a useful interface between the AOP framework and existing chemical screening and prioritization efforts by the US Environmental Protection Agency.
Keywords: Adverse Outcome Pathways (AOPs), risk assessment, data mining, database, data integration, association networks
Introduction
Efforts to inform toxicological risk assessment through the organization of biological information has led to the development of several pathway frameworks, such as the International Program on Chemical Safety (IPCS) mode-of-action (MOA) framework, and the National Research Council (NRC)-established toxicity pathway framework (Boobis et al., 2006; Boobis et al., 2008; Gibb, 2008). Most recently, the Organisation for Economic Co-operation and Development (OECD) has refined the concept of the adverse outcome pathway (AOP), a chemically agnostic framework that describes existing knowledge of biological perturbation in discrete steps across multiple scales of organization (Ankley et al., 2010; OECD, 2012). An AOP follows the events of a toxicity pathway in measurable mechanistic steps, beginning with a Molecular Initiating Event (MIE) that describes an action on a specific biomolecule, through a series of causally-linked Key Events (KE) representing downstream effects at the levels of molecular, cellular, tissue, organ, individual and population-level response, leading to some Adverse Outcome (AO).
To maximize the utility of the AOP construct, aggregation and standardized organization of AOP knowledge is essential. The OECD has developed a series of tools collectively known as the AOP Knowledge Base (AOP-KB) to provide a standardized, systematic structure for AOP development and knowledge dissemination. One such tool, the AOP-Wiki (www.aopwiki.org), facilitates collaborative AOP development by collecting and linking expert-curated AOP information through a controlled vocabulary (Villeneuve et al., 2014). This article introduces another tool for the development of AOPs: the AOP-DB is a database resource that relates the information stored in the AOP- Wiki to diseases, chemical-gene associations, taxonomic information, and other biological entities. These associations are sourced from public annotation and provide a wider context of relevant biology for each AOP, thus enabling researchers to expand predictions of chemical stressors and toxicological outcomes.
The AOP framework has already shown strong potential in toxicological chemical assessment, especially as a tool for the prioritization of chemicals, tissues, and organisms for effective assay design and the development of predictive toxicology models (OECD, 2012; Tollefsen et al., 2014). For example, the AOP can provide a basis for establishing structure-activity relationships between MIE molecular targets and specific xenobiotics of interest (Browne, Judson, Casey, Kleinstreuer, & Thomas, 2015; Landesmann, Goumenou, Munn, & Whelan, 2012; Vinken et al., 2013). The identification of intermediate KEs can inform in vitro assay endpoints (Vinken, 2013), which aids in the identification of toxicity pathways of interest. Further, defined realms of taxonomic applicability establish species context for regulatory decisions. Workflows to computationally predict taxonomic applicability for MIEs (LaLone et al., 2016) and AOPs (Mortensen, 2017; Mortensen, Pittman et al., in prep) have been described to predict species susceptibility and inform chemical evaluation. There is a wealth of free, publically available biological information on the web, including “omics” data, high throughput assays, and experimental and inferred associations between biological entities. However, the utility of this abundant knowledge is compromised by its vast dispersion over multiple formats and databases, requiring manipulation and extensive quality control to incorporate usable information. Multiple attempts have been made to integrate these resources in order to improve knowledge discovery, fill complementary information gaps, aid in data interpretation, and allow for collective mining of heterogeneous data types for a more comprehensive, systems-level view of biological questions (Gligorijevic & Przulj, 2015; Gomez-Cabrero et al., 2014; R. Judson et al., 2008; Lapatas, Stefanidakis, Jimenez, Via, & Schneider, 2015; Sherman et al., 2007; Smith et al., 2007; Wang, Khankhanian, Baranzini, & Mousavi, 2011). Integration of publically available data with the AOP framework has been explored for its utility in predicting putative AOPs. Incorporation of external data sources informs AOP discovery and development (Oki, Nelms, Bell, Mortensen, & Edwards, 2016).
To address the need for an automated system to relate AOPs to useful public information, we introduce the AOP-DB, a database resource that stores preliminary AOP-associated gene targets from the AOP-Wiki and relevant chemicals, diseases, biological pathways, taxonomy, and ontologies. The AOP-DB is a novel resource that integrates multiple heterogeneous data sources and types in a single centralized location, and automatically relates AOP mechanistic information to functional biological entities. The AOP-DB facilitates exploration across multiple levels of biological organization, and species-level variation. To enable the exchange of information among regulatory research efforts, the AOP-DB also interfaces between OECD AOP Discovery and Development tools, as well as and other EPA toxicology resources including ToxCast data (Dix et al., 2007) and the EPA Chemistry Dashboards (Richard & Williams, 2002). The AOP-DB seeks to capture the relationships between these diverse data types in order to profile AOPs in a systems biology context, act as a hypothesis-generation tool for assay development, and aid in the discovery of unidentified AOPs.
This paper briefly outlines the design and data sources of the AOP-DB, and illustrates the utility of the tool through three use cases: 1) characterizing the biological space of a putative AOP; 2) exploring the biological overlap between a collection of interconnected AOPs; and 3) hypothesis generation for the development of a potential AOP (e.g. an AOP not yet described). Here we illustrate the AOP-DB’s potential as a data integration tool for the characterization of AOPs, as well as a useful interface between OECD and EPA toxicology efforts and the wider landscape of publically available annotation. Though the AOP-DB is not currently available for public download, DB-sourced relationships between AOPs, KEs, chemical stressors, and ToxCast assays are integrated into the EPA Chemistry Dashboard (https://comptox.epa.gov/dashboard). Other database components are available upon request.
Materials and Methods
I. AOP-DB structure and quality validation
The goal of the AOP-DB is to capture relationships among genes and multiple biological entities to characterize AOP activity in a variety of contexts, such as associated diseases, chemicals, pathways, taxonomic groups, and protein interactions. The AOP-DB was developed on the SQL InnoDB platform version 5.6.36 (Oracle, 2000, 2013) and contains sixteen information tables linked through the centrally located gene identifier table, described below, and in Table 1 and Supplemental Figure 1. Quality control and assurance procedures used in the creation of the AOP-DB include data type validation, range validation, and enforced referential integrity, implemented as part of MySQL’s InnoDB storage engine format (Oracle, MySQL 5.7 Reference Manual, Accessed March 2017). These checks establish an expected data type (integer, string, etc.) and byte size for every column in the database, to ensure that entries are in the correct format and remain standardized across tables. Scripted routines were also included during the creation and update of database tables to ensure data integrity and consistency across tables, which is crucial in a denormalized database (Pinto, 2009) such as the AOP-DB. Since a denormalized database contains several instances of a single relationship in multiple tables (e.g. a gene appears with its associated taxon in both the disease-gene and chemical-gene table), table creation scripts programmatically ensure that these redundant relationships are updated concurrently with the same data to prevent inconsistencies across tables. Finally, sample queries with known expected results were designed to test the coherence and fidelity of the database, Table 1 gives the enumeration of entries for each information category in the database. Categories are described below:
Table 1.
Enumeration of each information type in the AOP-DB, with sources. The CTD chemical-gene associations, disease-gene associations, gene-gene interactions, and pairwise orthology scores are derived from secondary databases that combine multiple sources, in some cases calculating a confidence score. It should be noted that queries to these tables, especially for disease-gene and chemical-gene associations, often return large lists with spurious results due to the nature of the association derivation. For those tables that include confidence scores, it is recommended to use a threshold confidence score to filter out weaker association results, or to filter based on keywords to the biological question being asked.
| Biological category |
Data type | Count | Sources (Download Link) |
|---|---|---|---|
| Gene | Unique gene IDs: | 16808216 | NCBI Gene (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/) |
| Pairwise gene interaction scores: | 443295364 | STRING (http://string-db.org/cgi/download.pl) | |
| Taxonomy & orthology | Entrez-supported organisms: | 19292 | NCBI Taxonomy (ftp://ftp.ncbi.nlm.nih.gov/pub/HomoloGene/) |
| Orthologous groups: | 63138 | HomoloGene (ftp://ftp.ncbi.nlm.nih.gov/pub/) | |
| KEGG Orthology (http://www.kegg.jp/kegg/rest/keggapi.html) | |||
| Taxa supported by ortho groups: | 469 | ||
| Pairwise orthology confidence scores: | 40450671 | metaPhOrs (ftp://phylomedb.org/metaphors/) | |
| Taxa with pairwise confidence scores: | 1258 | ||
| AOP | Supported AOPs: | 103 | AOP-wiki (https://aopwiki.org/) |
| AOP-gene targets: | 564 | ||
| Chemical | CTD chemicals: | 164640 | CTD (http://ctdbase.org/downloads/) |
| CTD chemical-gene associations: | 846574 | ||
| DSSTox chemicals: | 22038 | AOP-wiki (https://aopwiki.org/) | |
| Direct DTX-AOP associations: | 188 | ||
| Inferred DTX-AOP associations: | 29768 | ||
| ToxCast assays: | 347 | ToxCast (ftp://newftp.epa.gov/comptox/High_Throughput_Screening_Data/Summary_Files) | |
| Inferred ToxCast assay-AOP links: | 353 | ||
| Pathway | Total pathways: | 75830 | KEGG Pathways (http://www.kegg.jp/kegg/rest/keggapi.html) |
| Pathway-entrez links: | 3663296 | Reactome (http://www.reactome.org/pages/download-data/) | |
| Inferred AOP-pathway associations: | 7340 | ConsensusPathDB (http://consensuspathdb.org/) | |
| Disease | Unique disease IDs: | 15093 | DisGeNET (http://www.disgenet.org/web/DisGeNET/menu/downloads) |
| Disease-gene associations: | 429036 | ||
| Inferred AOP-disease associations: | 33735 | ||
| Ontology | Unique GO Term IDs: | 25713 | NCBI Gene (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/) |
| Ontology-gene associations: | 1658739 |
A. AOP-gene associations
Associations between AOPs and their relevant gene IDs were mined from the AOP-Wiki based on ontology annotation ("https://aopwiki.org/," 2017; Ives, Campia, Wang, Wittwehr, & Edwards, in prep). Specifically, each KE is associated with an event component consisting of a Process, Object, Action, and Cellular or Organ Context ontology identifier; Object terms can be mapped to gene identifiers based on Protein Ontology (PR). Ontology annotation for all 715 KEs was downloaded from the AOP-Wiki, for all 164 AOPs as of December 2016. Of these total 164 AOPs, it is important to note that 113 are still considered “Under Development” by the OECD, and may be subject to change before they are formally endorsed.
B. Gene
The AOP-DB’s central linking entities are Entrez genes. The Entrez Gene database curated by the National Center for Biotechnology Information (NCBI) is made up of a set of unique identifiers for genes and corresponding gene-specific information, such as taxonomy, protein products, sequences, and orthologs (Maglott, Ostell, Pruitt, & Tatusova, 2011). These Entrez genes are sourced from fully-sequenced genomes of well-researched organisms. The AOP-DB hosts 1.68 million unique Entrez gene IDs to capture relationships between biological entities.
Entrez gene is not the only identification system for genes and proteins, and data sources occasionally use non-Entrez gene IDs. To ensure that the AOP-DB is capable of using these data as well, a built-in table is included for the mapping of alternative gene IDs to Entrez, curated by the Universal Protein Resource (UniProt, 2015).
C. Pathway
Biological pathways represent the series of molecular and genetic interactions that amount to the execution of a biological process. The AOP-DB makes use of three sources of pathway information: the Kyoto Encyclopedia of Genes and Genomes (KEGG), a database resource for the functional annotation and network organization of genes, accessed using the REST API (Kanehisa, Furumichi, Tanabe, Sato, & Morishima, 2017); Reactome, a database resource containing manually-curated pathway interactions sourced from PubMed literature (Croft et al., 2014); and Consensus PathDB, a composite database containing functional interaction data from 32 sources for human, mouse, and yeast pathways (Kamburov, Wierling, Lehrach, & Herwig, 2009). For each of these sources, pathways and their participating genes were downloaded and stored in the AOP-DB, along with outlinks to pathway visualizations where possible. The AOP-DB hosts 75,830 unique pathway IDs, with 3.7 million gene-pathway associations.
D. Chemical
The AOP-DB stores chemical-gene associations sourced from the Comparative Toxicogenomics Database (CTD), a resource that manually curates and infers relationships between genes, chemicals, and diseases from scientific literature (Davis et al., 2017). The AOP-DB hosts 164,640 chemicals from CTD, with 846,574 gene-chemical associations.
In addition to chemical-gene associations, the AOP-DB includes direct relationships between AOPs and chemical stressors in the EPA’s Distributed Structure-Searchable Toxicity (DSSTox) Database. DSSTox is a public resource that combines cheminformatics and toxicity data to aid in toxicology prediction and prioritization (Richard & Williams, 2002). By returning a list of DSSTox chemical IDs that are associated with an AOP, the AOP-DB makes it possible to interface AOP predictions and associations with the power of EPA’s iCSS Chemistry Dashboard (https://comptox.epa.gov/dashboard/). The AOP-DB hosts 22,038 chemicals from the DSSTox Database, with 188 confirmed DTX chemical-AOP associations mined directly from the AOP-Wiki, and approximately 30,000 inferred DTX chemical-AOP associations through CTD chemical-gene associations.
E. Disease
The associations between genes and human disease phenotypes in the AOP-DB are sourced from DisGeNET, which combines mined, curated, and inferred associations from ten sources for Mendelian, complex, environmental, and rare diseases as well as disease traits (Pinero et al., 2017). Due to the redundancy of information across these ten data sources, a confidence score between 0 and 1 was calculated for each association based on the proportion of the sources that recognize that association. The AOP-DB contains 15,093 unique disease outcomes with 429,036 gene-disease associations.
F. Gene orthology
Cross-species homology describes genes in two or more taxa descended from a common ancestor. Functional orthologs retain the same protein functions as the ancestral gene (Gabaldon & Koonin, 2013), allowing for the extrapolation of gene annotations across species based on ortholog concordance. In order to aid in the prediction of AOP susceptibility across different species, the AOP-DB includes orthology groupings from three sources: KEGG Orthology, NCBI HomoloGene, and the Meta-Phylogeny based Orthologs (metaPhOrs) database (Kanehisa et al., 2017; Pruitt & Maglott, 2001; Pryszcz, Huerta-Cepas, & Gabaldon, 2011). KEGG Orthology groups orthologous genes and proteins by their molecular functions, while NCBI HomoloGene creates groups from a BLASTp similarity-constructed taxonomy tree. The AOP-DB contains 63,138 orthologous gene groups among 469 taxa. In contrast, metaPhOrs combines seven phylogenetic tree databases to make pairwise orthology predictions for 4.1 million proteins. Due to the redundancy of information across and among the constituent data sources, a confidence score in a given orthology prediction can be calculated based on proportion of sources also making that prediction. The AOP-DB contains 40.4 million pairwise orthology confidence scores among 1,258 taxa.
G. Protein-protein Interactions
The AOP-DB returns gene-gene interaction scores sourced from the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING). The STRING database contains protein-protein interactions for over 2000 organisms, with each interaction scored by confidence level (Szklarczyk et al., 2015). Interactions are sourced from experimental data from primary databases, computational prediction from high-throughput experiments and orthology extrapolation, and text mining. The AOP-DB hosts 443 million pairwise gene interaction scores for 665,746 unique genes.
H. Gene Ontology
The Gene Ontology Consortium provides a controlled, machine-readable vocabulary that enables computational analysis of functional annotation for genes (Ashburner et al., 2000; Gene Ontology, 2015). In the AOP-DB, Entrez gene IDs are linked to their related GO biological process terms to help characterize the activity of a gene of interest. Currently, the AOP-DB contains 25,713 unique GO terms to describe the biological processes related to genes from 38 organisms.
I. ToxCast Assays
The US Environmental Protection Agency’s Toxicity ForeCaster (ToxCast™) program uses high-throughput in vitro assays to screen and prioritize chemicals for toxic effects (Dix et al., 2007; R. S. Judson et al., 2010). The AOP-DB stores the ToxCast in vitro assay information for 942 chemical-endpoint pairs and includes information about chemicals, targets, assay types, activity concentrations, and other pertinent metadata for analysis. Through the AOP-DB’s inferred associations, there are 432 ToxCast assays related to putative AOPs; these associations are fed to the EPA’s Chemistry Dashboard to inform researchers of existing pathways and data related to a chemical of interest.
II. AOP-DB Use Case Examples
Three use cases were chosen to demonstrate the utility of the AOP-DB: 1) characterizing the biological space of a putative AOP; 2) exploring the biological overlap between a collection of interconnected AOPs; and 3) hypothesis generation for the development of an AOP as-yet-undefined by the OECD. Output was visualized using Cytoscape network visualization software (Shannon et al., 2003).
Use Case 1: Characterization and Accuracy of Information
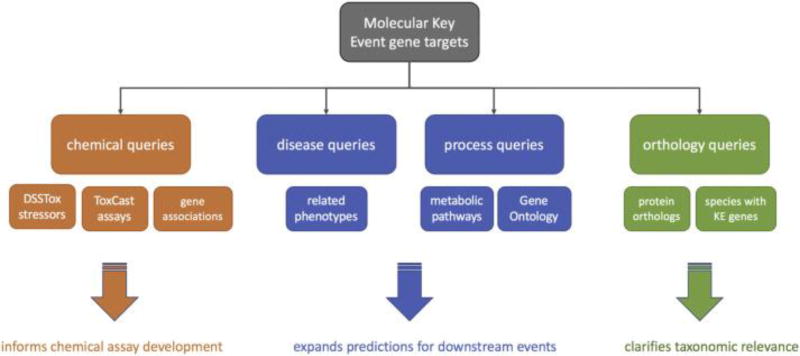
“Androgen Receptor (AR) Agonism Leading to Reproductive Dysfunction” is a putative AOP endorsed by the Extended Advisory Group on Molecular Screening and Toxicogenomics (EAGMST), which oversees the development and assessment of AOPs (https://aopwiki.org/aops/23, Accessed 15 March 2017). To demonstrate the accuracy of the information contained in the database and to showcase its utility as a hypothesis-generation tool, queries were constructed to return the AR agonism gene targets and their associations in the AOP-DB to return the chemical lists, biological pathways, and diseases associated with ARagonism and its downstream effects. A flow diagram of this procedure is shown in Figure 1A.
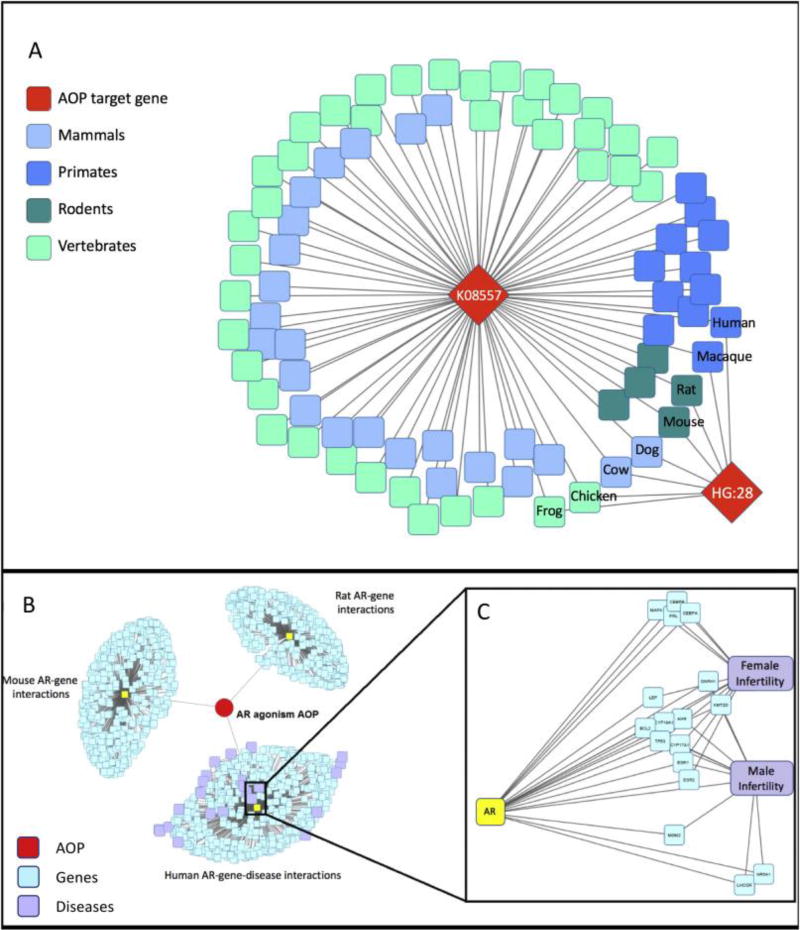
Figure 1.
Visualizations for AR agonism gene association data. 1A: Flowchart depicting the nature of potential lines of inquiry and their application. Queries constructed to examine the chemicals, diseases, functional processes, and species applicability of Key Evene genes are applied to assay development and prioritization efforts. 1B: The red diamond-shaped nodes represent the two orthology groups for the AR gene (ko:K08557 sourced from KEGG Orthology; HG:28 sourced from NCBI HomoloGene). Both of these sources recognize the model organisms H. sapiens, M. mulatta, M. musculus, R. norvegicus, C. lupus familiaris, B. taurus, X. tropicalis, and G. gallus as possessing orthologs for this gene. KEGG recognizes an additional 76 vertebrates with this gene. 1C: AR gene clustering based on STRING protein interaction confidence scores (visual cutoff = 0.5), and DisGeNET disease associations with those genes (visual cutoff = 0.1). The red circle and its edges represent the AR agonism AOP and its direct gene associations. The three large clusters represent the three taxa whose AOPwiki object IDs were associated with the AR gene identifier (in yellow): Human, Mouse, and Rat. Genes that are highly associated with AR (shown in blue) are clustered based on interaction confidence score. The human cluster (bottom) also shows gene-disease relationships for these closely-associated genes, with diseases represented by purple nodes. 1D: A subset of Network 1C, showing the genes that were both highly associated with the AR gene AND highly associated with the diseases “Female infertility” and “Male infertility”.
Use Case 2: Examining AOP Context
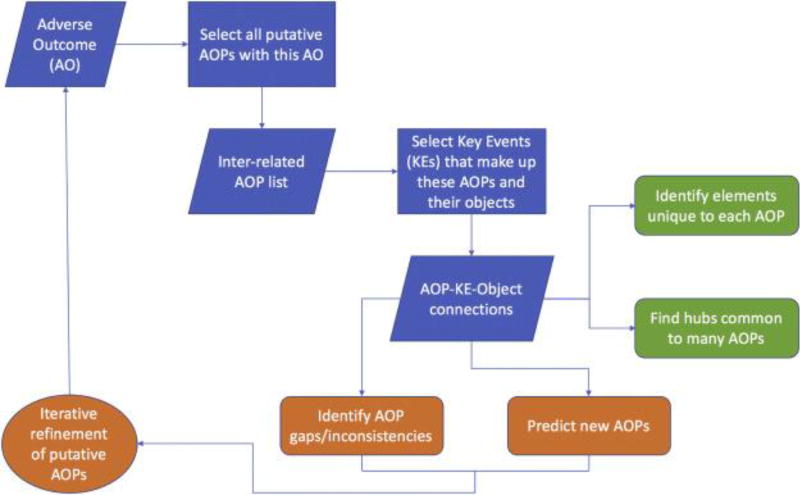
Of interest is the linking of related AOPs via shared KEs, in order to provide a wider biological context for potential downstream effects of a given chemical stressor or biological perturbation. A series of AOPs linked through shared KEs make it possible to computationally form AOP networks, which can supplement the creation of expert-curated AOPs and improve our systems-level understanding of how toxicity pathways proceed in a complex ecological context (Knapen, Vergauwen, Villeneuve, & Ankley, 2015). To explore the wider toxicological context of the AR agonism AOP, a group of related AOPs were selected by querying the AOP-DB for adverse outcomes involving “reproductive dysfunction.” The AOP-DB stores the controlled vocabulary used to describe the objects, actions, processes, and contexts of an AOP’s KEs, allowing users to automatically link AOPs in a network to examine their interactions. Results show the concordance and divergence between the reproductive dysfunction AOPs, identifying hub nodes (events and objects) that connect groups of closely-related reproductive toxicity pathways. A flow diagram for the examination and applications of putative AOP networks is shown in Figure 2A.
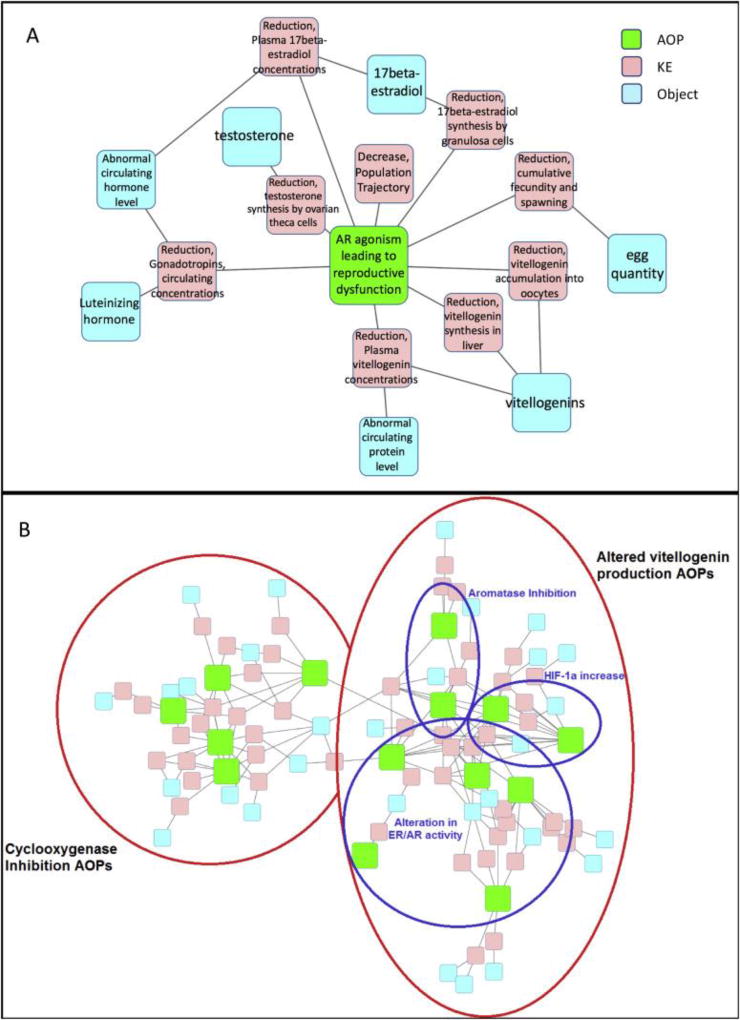
Figure 2.
Visual diagrams to show the relationships between AOPs, constituent KEs, and the objects upon which they act. 2A: Flowchart depicting the series of queries made to the AOP-DB to characterize the concordance and divergence between groups of AOPs. 2B: KE and object relationships for the AR agonism AOP alone, with the AOP shown in green, KE nodes shown in pink, and object nodes in blue. 2C: KE and object relationships for all “Reproductive Dysfunction” AOPs. The red circles represent the two major classes of reproductive dysfunction AOPs: Cyclooxygenase Inhibition AOPs (left), and altered Vitellogenin (VTG) production AOPs (right). Within the altered VTG class, there are three sub-classes of AOP types: those resulting from alteration in hormone receptor activity, those stemming from aromatase inhibition, and those resulting from an increase in HIF-1α concentration (demarcated by blue circles).
Use Case 3: Potential AOP Investigation and Hypothesis Generation
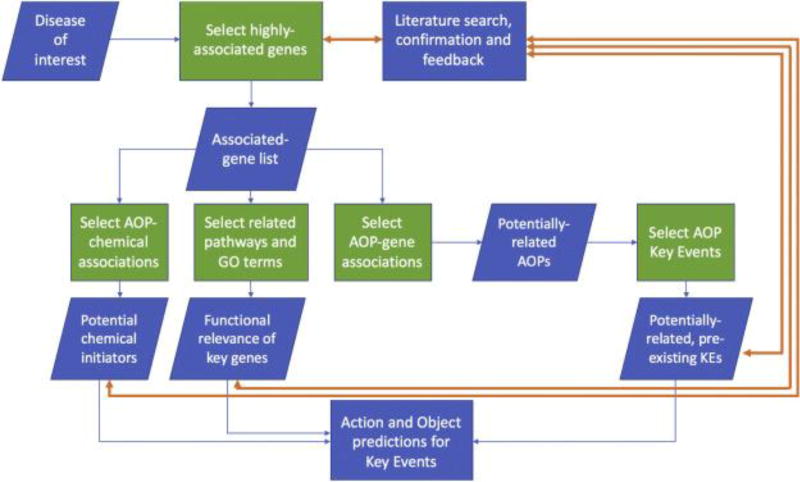
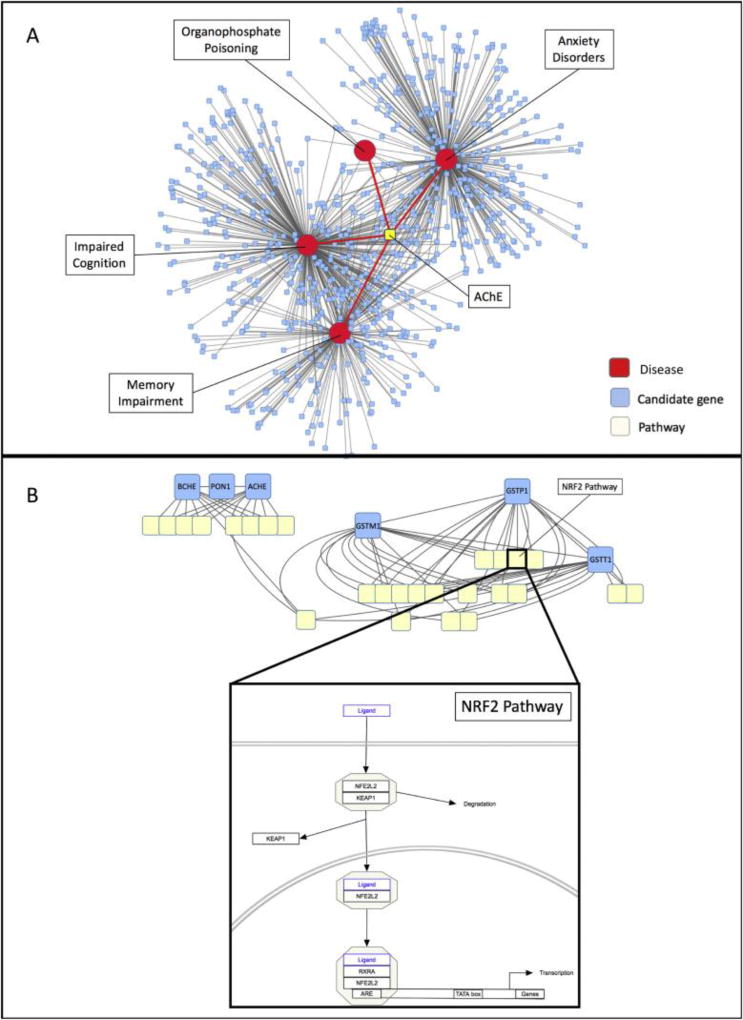
The AOP-DB may also be utilized to explore adverse outcomes that are not yet defined in the AOP framework. The AOP-DB can help to generate hypotheses for the identification of potential AOPs through a known outcome and its associated biological entities. The database was used to query associations across four diseases with closely-related gene associations (organophosphate poisoning, anxiety disorders, impaired cognition, and memory impairment), identify candidate KE genes, and explore the biological entities related to these candidate genes to characterize a potential AOP, “Acetylcholinesterase Inhibition Leading to Neuropsychological Dysfunction”. Figure 3A outlines a possible sequence of queries to characterize the biological activity of a potential AOP.
Figure 3.
Cytoscape visualizations for a proposed AOP, “acetylcholinesterase inhibition leading to neuropsychological dysfunction”. 3A: Flowchart depicting the series of steps taken to develop a predicted AOP from the AOP-DB. 3B: Four selected disease outcomes and their associated genes, clustered by gene interaction scores. Red nodes represent the diseases of interest, and blue nodes represent associated genes, clustered by association confidence score. 3C: Pathway membership for candidate genes in the predicted AOP “ACHE Inhibition leading to neuropsychological dysfunction.” Red nodes represent diseases of interest. Blue nodes represent the candidate genes associated with Organophosphate Poisoning and at least one of Anxiety Disorders, Memory Impairment, and Impaired Cognition. Each yellow node represents a pathway in which the indicated genes are active. 3D: Orthology confidence scores for gene candidates. The large light blue nodes are labelled with the identity of the gene candidate, and edges connect nodes of gene orthologs for different species based on confidence score. Edges connecting different genes (e.g. ACHE and BCHE genes) indicate paralogs (genes evolved after a duplication event).
Results
Database Summary & Quality Checks
Table 1 gives the enumeration and sources of entries for each information category in the database. The CTD chemical-gene associations, disease-gene associations, gene-gene interactions, and pairwise orthology scores are derived from secondary databases that combine multiple sources, and in some cases calculate a confidence score.
In addition to the datatype QA methods built into the MySQL database creation and population procedure, we also queried the database to demonstrate that the AOP-DB returns expected results in accordance with the information in the AOP-Wiki. Some sample queries to this effect are shown in Supplemental Figure 2. We find that the AOP-DB returns sensible results across all tables, showing ACHE-related biological entities by following related threads from DSSTox chemical-AOP associations, CTD chemical-gene-AOP associations, and disease-gene-AOP associations. The pathways and ontology terms related to this AOP return items such as “Cholinergic synapse” pathways and “acetylcholinesterase activity” biological processes, which are implicitly related to acetylcholinesterase function.
Use Case 1: Exploring and verifying the biological activity of a putative AOP – Androgen receptor agonism leading to reproductive dysfunction (Aop:23)
To summarize the AR agonism AOP in female teleost fish species, a chemical stressor binds to the androgen receptor and causes reduced testosterone and estrogen synthesis in the ovarian theca cells, eventually leading to a decrease in production of vitellogenin (egg-yolk protein) by females (Villeneuve, 2017). Established chemical initiators are comprised of non-aromatizable androgens, such as 17beta-trenbolone (Ankley et al., 2003).
We have queried the AOP-DB to show that the information stored within is consistent with the established literature, and provides avenues for further investigation. From the AOP gene targets stored in the AOP-Wiki, we returned the chemical lists, biological pathways, and diseases associated with androgen receptor agonism and its downstream effects (Supplemental Table 1). We confirm that the established chemical stressor trenbolone (Villeneuve, 2017) is implicated in this AOP through gene-target associations and identify additional potential chemical stressors such as metribolone and mibolerone, known AR agonists(Luderschmidt, Jawny, & Eiermann, 1987) (Hammond et al., 2001) (Table 2). This demonstrates that information in the AOP-DB is consistent with known chemical stressors listed for an AOP, and suggests additional chemicals potentially worthy of assay development. Through the AOP-DB’s gene orthology tables, we can examine which species and taxonomic divisions possess gene orthologs for the KE proteins necessary to the progression of the AOP (Figure 1B). For the AR agonism AOP, we see that vertebrate taxonomic groups across mammals, avians, reptiles, amphibians, and fish species all possess an ortholog for the AR; however, no invertebrate species were returned, suggesting that potential susceptibility of this AOP is limited to vertebrates.
Table 2.
An excerpt of query results for potentially-relevant chemicals to the androgen receptor agonism AOP. Trenbolone acetate, a form of the known AR agonist 17beta-trenbolone, is returned as a chemical that “binds to and results in increased activity of AR protein.” In addition, a host of other chemicals that act on the AR protein to increase its activity are also returned. For example, we see that the chemical metribolone (17beta-Hydroxy-17alpha-methylestra-4,9,11-trien-3-one, a synthetic non-aromatizable androgen) has many associations with AR and AR activity. Thus the information in the AOP-DB is consistent with known chemical stressors listed for an AOP, and suggests additional chemicals potentially worthy of assay development.
| MeSHID | Chemical Name | Interaction |
|---|---|---|
| C003600 | 11-ketotestosterone | 11-ketotestosterone binds to and results in increased activity of AR protein |
| C026486 | 1,2,5,6-dibenzanthracene | 1,2,5,6-dibenzanthracene binds to and results in increased activity of AR protein |
| C517232 | 1,2-dibromo-4-(1,2-dibromoethyl)cyclohexane | 1,2-dibromo-4-(1,2-dibromoethyl)cyclohexane binds to and results in increased activity of AR protein |
| C010422 | 2,4,6-trichlorophenyl 4-nitrophenyl ether | 2,4,6-trichlorophenyl 4-nitrophenyl ether binds to and results in increased activity of AR protein |
| C003524 | 4,16-androstadien-3-one | 4,16-androstadien-3-one analog binds to and results in increased activity of AR protein |
| D015741 | Metribolone | Metribolone binds to and results in increased activity of AR protein |
| C100075 | mibolerone | mibolerone binds to and results in increased activity of AR protein |
| D015058 | 1-Naphthylisothiocyanate | binds to and results in increased activity of AR |
| D009640 | Norethindrone | Norethindrone binds to and results in increased activity of AR protein |
| D009641 | Norethynodrel | Norethynodrel binds to and results in increased activity of AR protein |
| D016912 | Levonorgestrel | Levonorgestrel binds to and results in increased activity of AR protein |
| D053139 | Oseltamivir | Oseltamivir binds to and results in increased activity of AR protein |
| D010110 | Oxymetholone | Oxymetholone binds to and results in increased activity of AR protein |
| D011239 | Prednisolone | Prednisolone binds to and results in increased activity of AR protein |
| D011245 | Pregnadienes | Pregnadienes analog binds to and results in increased activity of AR protein |
| D011374 | Progesterone | Progesterone binds to and results in increased activity of AR protein |
| D013739 | Testosterone | Testosterone binds to and results in increased activity of AR protein |
| C479553 | tetrahydrogestrinone | tetrahydrogestrinone binds to and results in increased activity of AR protein |
| D014112 | Toxaphene | Toxaphene binds to and results in increased activity of AR protein |
| D014204 | Trenbolone Acetate | Trenbolone Acetate binds to and results in increased activity of AR protein |
| D014260 | Triclosan | Triclosan binds to and results in increased activity of AR protein |
By selecting KE genes, their closest gene interactions by confidence score, and the diseases most highly-associated with these genes, we can visually cluster the functional properties of an AOP. Figure 1C shows the clustering of protein-protein interactions based on confidence scores for the AR agonism KE genes, and includes disease associations with those genes. Filtering based on phenotypic outcome of interest, like diseases involving infertility, can provide predictions about the potential activity of an MIE in a wider context, identifying additional genes that may affect the progression of the AOP, as well as genes that participate in similar AOPs (Figure 1D).
Use Case 2: Examining overlap of a group of inter-related AOPs (AOPs with the “reproductive dysfunction” Adverse Outcome)
Figure 2B shows the KE and KE Object relationships for just the AR agonism AOP. By linking fourteen reproductive AOPs via KEs and object interactions shared between AOPs, we expand this view and distinguish between four major classes of reproductive dysfunction pathway initiators in the AOP-Wiki: cyclooxygenase inhibition, hormone receptor interference, aromatase interference, and HIF-1α-related dysfunctions (Figure 2C). The aromatase, AR/ER, and HIF-1α AOP groups all converge upon the KE “Vitellogenin accumulation into oocytes and oocyte growth/development, Reduction” and its downstream effects. Cyclooxygenase AOPs, on the other hand, lead to hormonal disruptions with a host of adverse outcomes (such as altered mating behavior and failed meiotic assembly checkpoints).
Use Case 3: Generating hypotheses for predicted AOPs – AChE Inhibition leading to Neuropsychological Dysfunction
Through casual exploration of disease links for proposed AOPs, we discovered that the gene targets involved in “Acetylcholinesterase (AChE) Inhibition Leading to Acute Mortality” (Russom, LaLone, Villeneuve, & Ankley, 2014) were also highly associated with negative neuropsychological effects, such as anxiety disorders (DisGeNET disease ID: umls:C0003469), impaired cognition (umls:C0338656), and memory impairment (umls:C0233794) (Figure 3B). Based on these associations, we predicted that AChE inhibition may also be associated with long-term neuropsychological dysfunction. In fact, this phenomenon is supported in the literature reference and is known as Chronic Organophosphate-Induced Neuropsychiatric Disorder (COPIND).
COPIND describes the group of co-occurring neuropsychological features that appear in occupational workers exposed to organophosphate pesticides, such as farm workers and sheep dippers (Ahmed & Davies, 2009). Whereas initial symptoms of acute organophosphate poisoning include neurophysiological effects like muscle paralysis and ataxia, follow-up studies of affected individuals have revealed behavioral changes including confusion, anxiety, depression, and problems with memory and concentration (Arun & Palimar, 2005). These symptoms persist for years after the reported initial exposures (Davies, Mrcpsych, Ahmed, & Freer, 2009; Institute of Medicine, 2000). Increased hippocampal ACh signaling in the brain and ACHE knockdown in the hippocampus lead to depression-like behaviors in mice, suggesting that ACHE inhibition (and the resulting accumulation of acetylcholine) could be the cause of these behaviors in individuals exposed to ACHE inhibitors like organophosphates (Mineur et al., 2013).
We selected genes that were shared between organophosphate poisoning and any of the other three examined disease outcomes. These genes are considered candidate Objects of a molecular KE (Table 3). From these candidate genes, we queried the AOP-DB to characterize the biological space of their collective activity, finding 66 pathways in which they participate (Figure 3C). These pathways can be examined for links to the disease outcomes of interest, such as “Cholinergic synapse” activity and “Neurotransmitter Clearance.” Thus the AOP-DB can suggest avenues of AOP progression through the linkage of MIEs to functional pathways. We also examine the orthology of the candidate genes, finding that ACHE and BCHE are likely paralogs, as well as GSTM1 and GSTP1, as each pair is closely phylogenetically linked (Figure 3D). We also see that all candidate genes show some level of relatedness to a wide variety of taxa, including invertebrates and fungi. However, the Adverse Outcome of neuropsychological dysfunction is implicitly limited to animal species for which behavior disturbance can be measured
Table 3.
Candidate genes for ACHE Inhibition Leading to Neuropsychological Dysfunction. These genes are those that were highly associated to Organophosphate poisoning and at least one of the following negative neuropsychological disease endpoints: anxiety disorders (DisGeNET disease ID: umls:C0003469), impaired cognition (umls:C0338656), and memory impairment (umls:C0233794).
| Entrez | Symbol | Gene name |
|---|---|---|
| 5444 | PON1 | Paraoxonase 1 |
| 43 | ACHE | Acetylcholinesterase |
| 2952 | GSTT1 | Glutathione S-transferase theta 1 |
| 2950 | GSTP1 | Glutathione S-transferase pi 1 |
| 590 | BCHE | Butyrylcholinesterase |
| 2944 | GSTM1 | Glutathione S-transferase mu 1 |
Discussion
We have presented the AOP-DB, a database resource that collects and connects public annotation information for chemicals, diseases, proteins, pathways, and their interactions through gene IDs to associate them with putative Adverse Outcome Pathways. The AOP-DB mines AOP-gene associations from the AOP-Wiki in a direct and automated fashion, allowing for the instantaneous integration of all available information for putative AOPs as they are developed. Important AOP associations, such as ToxCast assays relevant to putative AOPs and DSSTox chemical entries identified as AOP stressors, are then fed into other EPA computational toxicology tools to interface between AOP Discovery and Development efforts and existing data. Through queries constructed to follow entity-relationships across multiple tables, we confirm that the database returns consistent results for the characterization of the activity and context of putative AOPs (Supplemental Figure 2, Figure 1).
We predict that the data association types contained in the AOP-DB will be useful for researchers exploring the wider biological context of an AOP, for instance by: 1) providing expanded lists of genes potentially impacted by a given chemical stressor; 2) providing lists of genes that interact strongly with an AOP’s KE genes; 3) specifying potential disease outcomes of perturbations on KE genes, or genes of interest discovered through (1) and (2); 4) the intersection between an AOP of interest and all other putative AOPs defined in the AOP-Wiki, defined by shared KEs and Objects.
Based on MIEs tagged in the AOP-Wiki, chemical-gene associations can be used to identify groups of chemicals that might initiate a given AOP or expand a list of potential stressors, as with our first case study in which we identified the potential chemical stressors like mibolerone and metribolone, non-aromatizable androgens similar in structure to the established chemical stressor trenbolone (Table 2). Because the AOP-DB is focused on chemicals relevant to environmental toxicity, the database is especially useful for generating hypotheses regarding the action and impact of environmental chemical stressors.
Disease-gene associations can be used to recognize possible outcomes of perturbations on specific genes to generate hypotheses for potential AOPs, as with our case study predicting a potential AOP with the MIE “Acetylcholinesterase inhibition” and the AO “Neuropsychological dysfunction” (Figure 3C). Through exploration of the pathways linked to the disease-associated genes and confirmation through literature search, we named and identified the candidate KE “increased concentration of acetylcholine in the hippocampus,” leading to “increase in anxiety and depression-like symptoms.” Besides depression and anxiety, two organophosphate-related diseases were memory and cognitive impairment; however, we find that these sequelae require more evidence, or should perhaps comprise a separate adverse outcome, as we found no literature to support acetylcholine accumulation in the hippocampus directly leading to memory and cognitive dysfunction. Thus we show the AOP-DB’s utility in proposing potential AOPs, predicting constituent KEs, and identifying knowledge gaps that would benefit from additional research.
For a selected putative or computationally-predicted AOP, the AOP-DB can return a list of species that possess orthologs for all or some of those genes (Figure 1B). Use of such a result can provide a preliminary prediction for taxonomic applicability (e.g. if a taxonomic group lacks a functional ortholog for a gene that is fundamental to the progression of the MIE or downstream KEs, we then have information that these species are not at risk for that particular AOP, informing chemical prioritization). In addition, consistency scores for orthology predictions can give a measure of confidence that a given gene pair is a functional ortholog. Thus, the AOP-DB has the capability to illustrate, for a given list of genes, which species are predicted to have orthologs for these genes, and with what confidence. This feature makes the AOP-DB a useful tool for establishing a computational framework for defining a taxonomic range of applicability for an AOP. Currently, the AOP-DB’s AOP-gene, gene-pathway, and gene-orthology relationships are being combined to examine and compare the systems-level activity of AOP-related genes across organisms as part of a computational workflow to define taxonomic applicability for AOPs (Mortensen, Pittman et al., in prep). Note, however, that the confidence scores for orthology prediction are derived from consistency of annotation across several sources and not on physical similarity; therefore, some members of more-closely related taxonomic groups may show lower confidence scores than more distantly-related groups (see Figure 3D for examples of this phenomenon). In the future, we hope to include measures of protein similarity instead of annotation consistency.
The ontology terms tagged in the AOPwiki and stored in the AOP-DB can link a group of interrelated AOPs by shared KEs and objects, expanding predictive toxicity models and supporting the development of related AOPs. Reproductive dysfunction networks have been previously investigated by (Knapen et al. (2015)), in which multiple AOPs were linked via altered vitellogenin (egg-yolk protein) mRNA expression. Our AOP network constructed from the putative reproductive dysfunction AOPs is consistent with their finding that aromatase and AR/ER interference converge upon altered vitellogenin production; our methods also include HIF-1α AOPs in this group, as well as a separate class of reproductive AOPs initiated by Cyclooxygenase inhibition (Figure 2C). While previous methods used information curated manually from the AOP-Wiki, the AOP-DB allows for fast, automatic creation of these networks, thus dramatically expediting such efforts to aid in predictive toxicology.
The AOP-DB is uniquely useful as a single integrated source of biological information that is interconnected with AOP knowledge. We foresee the AOP-DB being used by researchers using the AOP-DB to elaborate upon their chemical and gene target hypotheses using association tables, or to elucidate mechanisms through pathway, gene interaction, and KE information tables. Many of the AOP-DB association tables cover a substantial amount of experimental and secondary association data, which can lead to the return of spurious results. Though confidence scores can provide a primary filter, this strategy comes with caveats. Confidence scores, for example those leveraged by the gene-orthology, gene-disease, and gene-interaction associations, do not necessarily coincide with AOP-relevance. For AOPs that are not well-characterized in the scientific literature, associations may be mentioned by few sources, and thus return a relatively low confidence score compared to well-studied associations. While network computational analysis would be an interesting avenue to explore in the AOP-DB, prioritized ranking based on confidence scores alone would not necessarily converge with the biological evidence supporting an AOP of interest. Therefore, at present we recommend that AOP-DB analyses be accompanied by the interpretation of skilled toxicologists and biologists to identify likely candidates for further pathway exploration and to assess the implications of AOP relationships.
The AOP-DB can therefore be considered as an AOP profiling and exploration tool that gives a broad, systems-level overview of the context in which putative AOPs operate. The AOP-DB simplifies the creation of network visualizations, using data points as nodes and association scores as edges, which will help researchers view and understand the action of KEs not only in the systems context of general biology, but also in the context of existing AOP vocabulary. Since one of the goals of the AOP structure is to use standardized language with a reusable “building block” philosophy for KEs and their relationships, the AOP-DB is especially suited for proposing new AOPs related to existing entries.
It should be noted that some of the AOP-DB’s parent IDs have no association data; for example, though the AOP-DB contains entries for 16.8 million Entrez genes, only 2.1 million of those gene IDs have data for protein interaction scores. The AOP-DB stores all available gene, chemical, disease, etc. IDs from its constituent sources, regardless of whether these IDs have data for all association types. As new sources are added and existing sources are updated, we hope that these data gaps will be filled. In the meantime, orphan IDs and associations will remain to provide what information they can for hypothesis generation. Another caveat is that the AOP-DB stores the information found in its constituent sources without skepticism or alteration. Though we have ensured that all the sources from which the AOP-DB draws its information use robust and peer-reviewed methods, the false positives and false negatives native to these sources are transferred to the AOP-DB. However, we predict that our use of multiple sources for certain datatypes may offset some false negatives.
Finally, the relationships between AOPs and their KEs, objects, and chemical stressors are limited to those that have been defined in the AOP-Wiki. Some entries are still under development and may be subject to change before formal endorsement. These gaps and uncertainties will be resolved as the AOP-KB grows and matures, but in the meantime it should be noted that AOP discovery and development is in its preliminary stages, and by extension the information contained in the AOP-DB is under development.
In summary, the AOP-DB provides an interface between chemical prioritization efforts and the AOP-KB, and automatically integrates large-scale gene association data for the characterization of putative and predicted AOPs in a variety of contexts. Since the AOP-DB is standardized by gene linkages, future integration of new data sources and types is as simple as finding Entrez gene associations with the relevant data points. Because the AOP-DB leverages gene-gene association scores to cluster gene lists by confidence-of-interaction level, one future use could be to computationally prioritize related gene interactions, and thus predict the downstream molecular effects of an MIE binding. Another potential use of the AOP-DB is to screen putative AOPs for disease and chemical association in order to characterize human population level genetic susceptibility (Mortensen, Chamberlin, et al., in prep), which directly contributes to the EPAs explicit consideration of vulnerable populations as laid out in the Frank R. Lautenberg Chemical Safety for the 21st Century Act to reform the Toxic Substances Control Act.
Supplementary Material
Supplemental Figure 1: Entity-relationship diagram for the AOP-DB, showing the relationships between each data type. Though most entities are related via associations with Entrez genes, there also exists a series of tables for the association of AOPs to known chemical stressors from the DSSTox Dashboard (AOP_stressor and stressor_info, bottom left corner).
Supplemental Figure 2: Sample AOP-DB queries and results, following the associations stored for the “Acetylcholinesterase (ACHE) inhibition leading to acute mortality” AOP. Key rows emphasizing the concordance of information are highlighted in yellow. Related threads can be followed from DSSTox chemical-AOP associations, CTD chemical-gene-AOP associations, and disease-gene-AOP associations. The pathways and ontology terms related to this AOP return sensible, obviously-related results, such as “Cholinergic synapse” pathways and “acetylcholinesterase activity” biological processes.
Supplemental Table 1A: Pathways associated with the AR agonism AOP. The gene target list was first expanded through AOP-DB orthology, since pathway annotation for fish taxa of established relevance is sparse. Pathway exploration could be useful in the development of AOPs in which the precise series of molecular interactions is unknown, as pathways illustrate the reactions by which systems-level processes unfold. For this reason, the AOP-DB provides web addresses to the source network visualizations where possible.
Supplemental Table 1B: Diseases associated with the AR agonism AOP, sorted by confidence score (cutoff = 0.1). The AOP-DB finds associations with diseases that one might expect to see for the AR gene target, including the AOP, female infertility. Note that the confidence score is simply based on the number of sources postulating the gene-disease association, and does not indicate AOP-relevance. A list of associated diseases can help researchers to hypothesize new Adverse Outcomes related to chemical stressors and genes under investigation. For example, what interactions with the AR lead to “malignant neoplasms of prostate” and “mammary neoplasms”? Could those disease pathways be activated by a specific MIE and described in the AOP framework?
Acknowledgments
The information in this document has been funded wholly by the U. S. Environmental Protection Agency, and supports the Agency’s Office of Research and Development’s Chemical Safety and Sustainability Research Action Plan Project 12.01: Adverse Outcome Pathway Discovery and Development, Task Number 1.1c: Taxonomic Relevance of AOPs. This project follows the Quality Assurance Project Protocol described in: IRP-NHEERL-RTP/RCU/GRC/HM /2016-01-r1, and the Operating Procedures described in: NHEERL/RCU/HMM/2016-05-r0.This document has been subjected to review by the National Health and Environmental Effects Research Laboratory and approved for publication. Approval does not signify that the contents reflect the views of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use. We thank Chris Grulke, Jeff Edwards, and Antony Williams for their work in integrating the AOP-DB and the DSSTox Chemistry Dashboards. We would also like to thank Dr. Ronald Hines, Dr. Russell Owen, Dr. Katherine Dionisio, and Dr. Michelle Angrish for their thoughtful reviews and suggestions for improvement of the manuscript.
Abbreviations
- ACHE
acetylcholinesterase
- ACh
acetylcholine
- AO
Adverse Outcome
- AOP
Adverse Outcome Pathway
- AOP-DB
Adverse Outcome Pathway Data Base
- AOP-KB
Adverse Outcome Pathway Knowledge Base
- AR
Androgen Receptor
- COPIND
Chronic OrganoPhosphate-Induced Neuropsychiatric Disorder
- CTD
Comparative Toxicogenomics Database
- DSSTox
Distributed Structure-Searchable Toxicity
- EAGMST
Extended Advisory Group on Molecular Screening and Toxicogenomics
- ER
Estrogen Receptor
- HIF-1α
Hypoxia-inducible factor 1-alpha
- iCSS
Interactive Chemical Safety for Sustainability
- IPCS
International Programme on Chemical Safety
- KE
Key Event
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MIE
Molecular Initiating Event
- MOA
Mode of Action
- NCBI
National Center for Biotechnology Information
- NRC
National Research Council
- OECD
Organisation for Economic Co-operation and Development
- QA
Quality Assurance
- REST API
Representational State Transfer Application Program Interface
- ToxCast
Toxicity Forecaster
Footnotes
Author Contributions
H.M.M. conceived the project. H.M.M. and M.P. designed the database and selected data sources. M.P. wrote code to upload and integrate data sources, and construct and populate the database. M.P. and C.I. performed ontology mapping. M.P. and S.E. wrote code to integrate the database with AOPwiki and CSS dashboards. M.P. and H.M.M. wrote the manuscript. All authors reviewed, revised and provided feedback on the manuscript.
Works Cited
- Ahmed GM, Davies DR. Chronic organophosphate exposure: toward the definition of a neuropsychiatric syndrome. Journal of Nutritional and Environmental Medicine. 2009;7:169–176. doi: 10.1080/13590849762583. [DOI] [Google Scholar]
- Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, Villeneuve DL. Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ Toxicol Chem. 2010;29(3):730–741. doi: 10.1002/etc.34. [DOI] [PubMed] [Google Scholar]
- Ankley GT, Jensen KM, Makynen EA, Kahl MD, Korte JJ, Hornung MW, Gray LE. Effects of the androgenic growth promoter 17-beta-trenbolone on fecundity and reproductive endocrinology of the fathead minnow. Environ Toxicol Chem. 2003;22(6):1350–1360. [PubMed] [Google Scholar]
- Arun M, Palimar V. Neurological manifestations in Organophosphorous toxicity. Journal of Indian Academic Forensic Medicine. 2005;30:29–31. [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boobis AR, Cohen SM, Dellarco V, McGregor D, Meek ME, Vickers C, Farland W. IPCS framework for analyzing the relevance of a cancer mode of action for humans. Crit Rev Toxicol. 2006;36(10):781–792. doi: 10.1080/10408440600977677. [DOI] [PubMed] [Google Scholar]
- Boobis AR, Doe JE, Heinrich-Hirsch B, Meek ME, Munn S, Ruchirawat M, Vickers C. IPCS framework for analyzing the relevance of a noncancer mode of action for humans. Crit Rev Toxicol. 2008;38(2):87–96. doi: 10.1080/10408440701749421. [DOI] [PubMed] [Google Scholar]
- Browne P, Judson RS, Casey WM, Kleinstreuer NC, Thomas RS. Screening Chemicals for Estrogen Receptor Bioactivity Using a Computational Model. Environ Sci Technol. 2015;49(14):8804–8814. doi: 10.1021/acs.est.5b02641. [DOI] [PubMed] [Google Scholar]
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, D'Eustachio P. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–477. doi: 10.1093/nar/gkt1102. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DR, Mrcpsych M, Ahmed GM, Freer T. Organophosphate Induced Neuropsychiatric Disorder (COPIND): Results of Two Postal Questionnaire Surveys. Journal of Nutritional and Environmental Medicine. 2009;9(2):123–134. doi: 10.1080/13590849961726. [DOI] [Google Scholar]
- Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, Mattingly CJ. The Comparative Toxicogenomics Database: update 2017. Nucleic Acids Res. 2017;45(D1):D972–D978. doi: 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, Kavlock RJ. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol Sci. 2007;95(1):5–12. doi: 10.1093/toxsci/kfl103. [DOI] [PubMed] [Google Scholar]
- Gabaldon T, Koonin EV. Functional and evolutionary implications of gene orthology. Nat Rev Genet. 2013;14(5):360–366. doi: 10.1038/nrg3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–1056. doi: 10.1093/nar/gku1179. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb S. Toxicity testing in the 21st century: a vision and a strategy. Reprod Toxicol. 2008;25(1):136–138. doi: 10.1016/j.reprotox.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Gligorijevic V, Przulj N. Methods for biological data integration: perspectives and challenges. J R Soc Interface. 2015;12(112) doi: 10.1098/rsif.2015.0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cabrero D, Abugessaisa I, Maier D, Teschendorff A, Merkenschlager M, Gisel A, Tegner J. Data integration in the era of omics: current and future challenges. BMC Syst Biol. 2014;8(Suppl 2):I1. doi: 10.1186/1752-0509-8-S2-I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J, Le Q, Goodyer C, Gelfand M, Trifiro M, LeBlanc A. Testosterone-mediated neuroprotection through the androgen receptor in human primary neurons. J Neurochem. 2001;77(5):1319–1326. doi: 10.1046/j.1471-4159.2001.00345.x. [DOI] [PubMed] [Google Scholar]
- https://aopwiki.org/ [Retrieved March 25, 2017];Wiki 2.0 Upgrade. 2017 Dec 4; 2016 from https://aopwiki.org/
- Fulco CE, Liverman CT, Sox HC, editors. Institute of Medicine. Gulf War and Health: Volume 1. Depleted Uranium, Sarin, Pyridostigmine Bromide, Vaccines. Washington (DC): 2000. [PubMed] [Google Scholar]
- Ives C, Campia I, Wang R-L, Wittwehr C, Edwards S. Creating a Structured AOP Knowledgebase via Ontology-Based Annotations. doi: 10.1089/aivt.2017.0017. in prep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R, Richard A, Dix D, Houck K, Elloumi F, Martin M, Wolf M. ACToR--Aggregated Computational Toxicology Resource. Toxicol Appl Pharmacol. 2008;233(1):7–13. doi: 10.1016/j.taap.2007.12.037. [DOI] [PubMed] [Google Scholar]
- Judson RS, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, Mortensen HM, Dix DJ. In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project. Environ Health Perspect. 2010;118(4):485–492. doi: 10.1289/ehp.0901392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB--a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–628. doi: 10.1093/nar/gkn698. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapen D, Vergauwen L, Villeneuve DL, Ankley GT. The potential of AOP networks for reproductive and developmental toxicity assay development. Reprod Toxicol. 2015;56:52–55. doi: 10.1016/j.reprotox.2015.04.003. [DOI] [PubMed] [Google Scholar]
- LaLone CA, Villeneuve DL, Lyons D, Helgen HW, Robinson SL, Swintek JA, Ankley GT. Editor's Highlight: Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS): A Web-Based Tool for Addressing the Challenges of Cross-Species Extrapolation of Chemical Toxicity. Toxicol Sci. 2016;153(2):228–245. doi: 10.1093/toxsci/kfw119. [DOI] [PubMed] [Google Scholar]
- Landesmann B, Goumenou M, Munn S, Whelan M. In: Description of Prototype Modes-of-Action Related to Repeated Dose Toxicity. J. S. a. P. Reports, editor. European Commission Joint Research Center Institute for Health and Consumer Protection; 2012. [Google Scholar]
- Lapatas V, Stefanidakis M, Jimenez RC, Via A, Schneider MV. Data integration in biological research: an overview. J Biol Res (Thessalon) 2015;22(1):9. doi: 10.1186/s40709-015-0032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderschmidt C, Jawny J, Eiermann W. Relative binding affinity at metribolone androgenic binding sites of various antiandrogenic agents. Arzneimittelforschung. 1987;37(11):1262–1265. [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–57. doi: 10.1093/nar/gkq1237. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineur YS, Obayemi A, Wigestrand MB, Fote GM, Calarco CA, Li AM, Picciotto MR. Cholinergic signaling in the hippocampus regulates social stress resilience and anxiety- and depression-like behavior. Proc Natl Acad Sci U S A. 2013;110(9):3573–3578. doi: 10.1073/pnas.1219731110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen H. Defining a Computational Framework for the Assessment of Taxonomic Applicability. The Toxicologist, Supplement to the Toxicological Sciences. 2017;1(156):88. [Google Scholar]
- Mortensen H, Pittman M, LaLone CA, Edwards S, Villeneuve DL, Ankley GT. A Computational Framework for Defining the Taxonomic Applicability of the Adverse Outcome Pathway. (in prep) [Google Scholar]
- Mortensen H, Chamberlin J, Angrish M, Lee J, Joubert B, Sipes N, Euling S. Defining Human Genetic Inter-Individual Variability in 21st Century Risk Assessment, Mammalian Genome Special Issue--Genetics & Environment. (in prep) [Google Scholar]
- OECD. Series on Testing and Assessment. Organisation for Economic Co-operation and Development (OECD); 2012. The Adverse Outcome Pathway for Skin Sensitisation Initiated by Covalent Binding to Proteins, Part 1: Scientific Evidence. [Google Scholar]
- Oki NO, Nelms MD, Bell SM, Mortensen HM, Edwards SW. Accelerating Adverse Outcome Pathway Development Using Publicly Available Data Sources. Curr Environ Health Rep. 2016;3(1):53–63. doi: 10.1007/s40572-016-0079-y. [DOI] [PubMed] [Google Scholar]
- Oracle. [Retrieved March 2017];MySQL 5.7 Reference Manual: The InnoDB Storage Engine. from https://dev.mysql.com/doc/refman/5.7/en/innodb-storage-engine.html.
- Oracle. MySQL InnoDB (Version 5.6.36) 2000, 2013 [Google Scholar]
- Pinero J, Bravo A, Queralt-Rosinach N, Gutierrez-Sacristan A, Deu-Pons J, Centeno E, Furlong LI. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto Y. A Framework for Systematic Database Denormalization. Global Journal of Computer Science and Technology. 2009;(9):44–52. [Google Scholar]
- Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29(1):137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryszcz LP, Huerta-Cepas J, Gabaldon T. MetaPhOrs: orthology and paralogy predictions from multiple phylogenetic evidence using a consistency-based confidence score. Nucleic Acids Res. 2011;39(5):e32. doi: 10.1093/nar/gkq953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard AM, Williams CR. Distributed structure-searchable toxicity (DSSTox) public database network: a proposal. Mutat Res. 2002;499(1):27–52. doi: 10.1016/s0027-5107(01)00289-5. [DOI] [PubMed] [Google Scholar]
- Russom CL, LaLone CA, Villeneuve DL, Ankley GT. Development of an adverse outcome pathway for acetylcholinesterase inhibition leading to acute mortality. Environ Toxicol Chem. 2014;33(10):2157–2169. doi: 10.1002/etc.2662. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman BT, Huang da W, Tan Q, Guo Y, Bour S, Liu D, Lempicki RA. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Lewis S. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefsen KE, Scholz S, Cronin MT, Edwards SW, de Knecht J, Crofton K, Patlewicz G. Applying Adverse Outcome Pathways (AOPs) to support Integrated Approaches to Testing and Assessment (IATA) Regul Toxicol Pharmacol. 2014;70(3):629–640. doi: 10.1016/j.yrtph.2014.09.009. [DOI] [PubMed] [Google Scholar]
- UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–212. doi: 10.1093/nar/gku989. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve DL. [Retrieved March 25, 2017];Androgen receptor agonism leading to reproductive dysfunction. 2017 Mar 20; 2017 from https://aopwiki.org/aops/23.
- Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, Whelan M. Adverse outcome pathway (AOP) development I: strategies and principles. Toxicol Sci. 2014;142(2):312–320. doi: 10.1093/toxsci/kfu199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinken M. The adverse outcome pathway concept: a pragmatic tool in toxicology. Toxicology. 2013;312:158–165. doi: 10.1016/j.tox.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Vinken M, Landesmann B, Goumenou M, Vinken S, Shah I, Jaeschke H, Rogiers V. Development of an adverse outcome pathway from drug-mediated bile salt export pump inhibition to cholestatic liver injury. Toxicol Sci. 2013;136(1):97–106. doi: 10.1093/toxsci/kft1777. [DOI] [PubMed] [Google Scholar]
- Wang L, Khankhanian P, Baranzini SE, Mousavi P. iCTNet: a Cytoscape plugin to produce and analyze integrative complex traits networks. BMC Bioinformatics. 2011;12:380. doi: 10.1186/1471-2105-12-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Entity-relationship diagram for the AOP-DB, showing the relationships between each data type. Though most entities are related via associations with Entrez genes, there also exists a series of tables for the association of AOPs to known chemical stressors from the DSSTox Dashboard (AOP_stressor and stressor_info, bottom left corner).
Supplemental Figure 2: Sample AOP-DB queries and results, following the associations stored for the “Acetylcholinesterase (ACHE) inhibition leading to acute mortality” AOP. Key rows emphasizing the concordance of information are highlighted in yellow. Related threads can be followed from DSSTox chemical-AOP associations, CTD chemical-gene-AOP associations, and disease-gene-AOP associations. The pathways and ontology terms related to this AOP return sensible, obviously-related results, such as “Cholinergic synapse” pathways and “acetylcholinesterase activity” biological processes.
Supplemental Table 1A: Pathways associated with the AR agonism AOP. The gene target list was first expanded through AOP-DB orthology, since pathway annotation for fish taxa of established relevance is sparse. Pathway exploration could be useful in the development of AOPs in which the precise series of molecular interactions is unknown, as pathways illustrate the reactions by which systems-level processes unfold. For this reason, the AOP-DB provides web addresses to the source network visualizations where possible.
Supplemental Table 1B: Diseases associated with the AR agonism AOP, sorted by confidence score (cutoff = 0.1). The AOP-DB finds associations with diseases that one might expect to see for the AR gene target, including the AOP, female infertility. Note that the confidence score is simply based on the number of sources postulating the gene-disease association, and does not indicate AOP-relevance. A list of associated diseases can help researchers to hypothesize new Adverse Outcomes related to chemical stressors and genes under investigation. For example, what interactions with the AR lead to “malignant neoplasms of prostate” and “mammary neoplasms”? Could those disease pathways be activated by a specific MIE and described in the AOP framework?