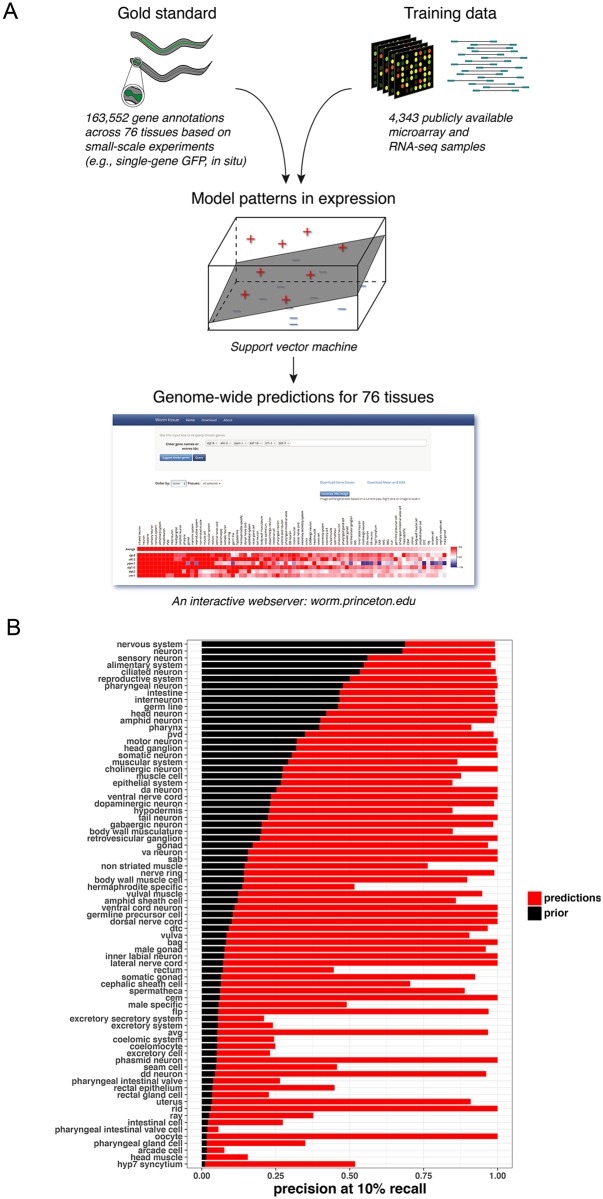
Fig 5. New prediction tool for sub-tissue/cellular gene expression.
A) Overview of the tissue expression prediction tool. We obtained 163,552 curated gene annotations to tissue expression based on small-scale experiments from WormBase and the C. elegans Tissue Expression Consortium (gold standard) and collected 4,372 microarray and RNA-seq samples (training data). For each of 76 tissues and cell types, we integrated the gold standard and training data using a support vector machine to make genome-wide predictions. Finally, we built an interactive webserver (worm.princeton.edu), where users can explore and download these predictions. B) Prediction performance of tissue expression models. Precision at 10% recall based on 5-fold cross-validation is shown. High precision is achieved even for tissues and cell types where there are relatively few known examples of tissue-specific expression based on small-scale experiments (low prior, where prior is the expected fraction of positive predictions if genes were chosen at random).