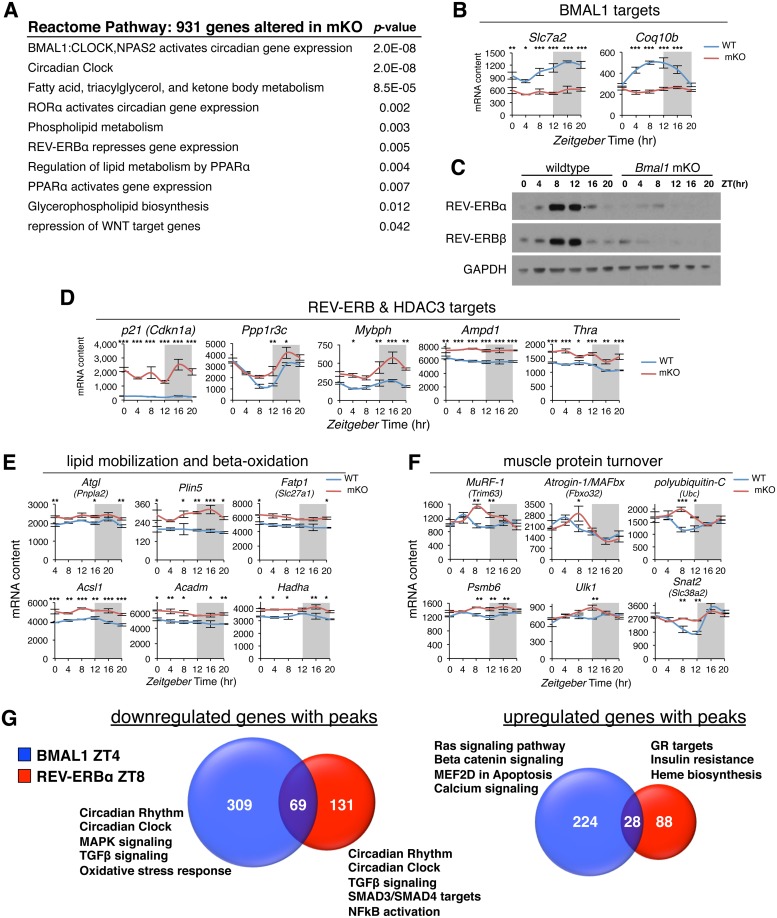
Fig 2. Transcriptional reprogramming of metabolic pathways in mKO muscles.
(A) REACTOME pathway enrichment analysis of 931 differentially expressed genes from mKO TA muscles. (B) Diurnal expression profiles of selected BMAL1-dependent circadian genes in TA determined by microarray and plotted as absolute expression levels (n = 3 × timepoint; mean ± SEM *p = 0.05, **p = 0.01, ***p = 0.001, 2-way ANOVA with Bonferroni correction). (C) Diurnal REV-ERBα and REV-ERBβ protein levels determined by western blot in vastus lateralis; GAPDH used as loading control. (D-F) Diurnal expression profiles in TA muscle of selected (D) REV-ERBα and HDAC3 target genes, (E) PPARα/δ-regulated mediators of lipid catabolism and oxidation, (F) GR-regulated mediators of protein turnover (n = 3 × timepoint; mean ± SEM *p = 0.05, **p = 0.01, ***p = 0.001, 2-way ANOVA with Bonferroni correction). (G) Venn diagram showing relative overlap and pathway enrichment of differentially regulated mKO genes with BMAL1 and REV-ERBα ChIP-seq peaks. Underlying data can be found in supporting files S1 Data, S1 Table, and at Gene Expression Omnibus (accession number GSE43071). BMAL1, brain and muscle ARNT-like protein 1; ChIP-seq, chromatin immunoprecipitation followed by next-generation sequencing; CLOCK, circadian locomotor output cycles kaput; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GR, glucocorticoid receptor; HDAC3, histone deacetylase 3; MAPK, mitogen-activated protein kinase; MEF2, myocyte enhancer binding factor 2; mKO, myocyte-specific loss of BMAL1; NFκB, nuclear factor kappa B; NPAS2, neuronal PAS domain protein 2; PPARα/δ, peroxisome proliferator–activated receptor alpha/delta; RORα, RAR-related orphan receptor alpha; TA, tibialis anterior; TGFβ, transforming growth factor beta; WT, wild type; ZT, Zeitgeber time.