Fig. 1. Acute sleep loss induces changes in DNA methylation in adipose tissue in healthy humans.
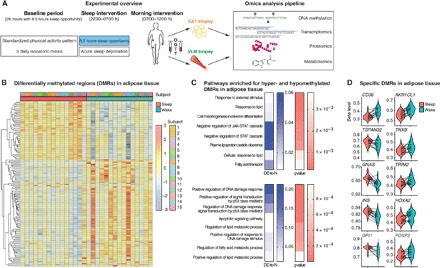
(A) Participants were investigated both after a night of sleep loss (that is, overnight wakefulness) and after a night of normal sleep, in each condition after an in-lab baseline day and night (26 hours in total, with an 8.5-hour baseline sleep opportunity) with standardized physical activity levels and isocaloric meals. Biopsies from the vastus lateralis muscle (VLM) and subcutaneous adipose tissue (SAT), as well as fasting blood sampling, preceded an oral glucose tolerance test (OGTT) and subsequent blood sampling. This was followed by a pipeline of omic analyses across tissues. (B) Differentially methylated regions (DMRs; FDR < 0.05) in adipose tissue showing DNA methylation (beta levels) after sleep and sleep loss (wake) across the 15 participants, with hierarchical clustering of DMR beta levels (z scores). (C) Significant gene ontology (GO) annotations based on hypermethylated (top) and hypomethylated DMRs (bottom) in adipose tissue in response to sleep loss, showing the ratio of differentially expressed gene-associated DMRs (DE) to the total number (N) of genes in a given pathway (“DE-to-N”) and adjusted P values (q values, FDR< 0.05). (D) Beta levels across some of the most significant DMRs in adipose tissue, in proximity to the specified genes, following sleep and sleep loss.
