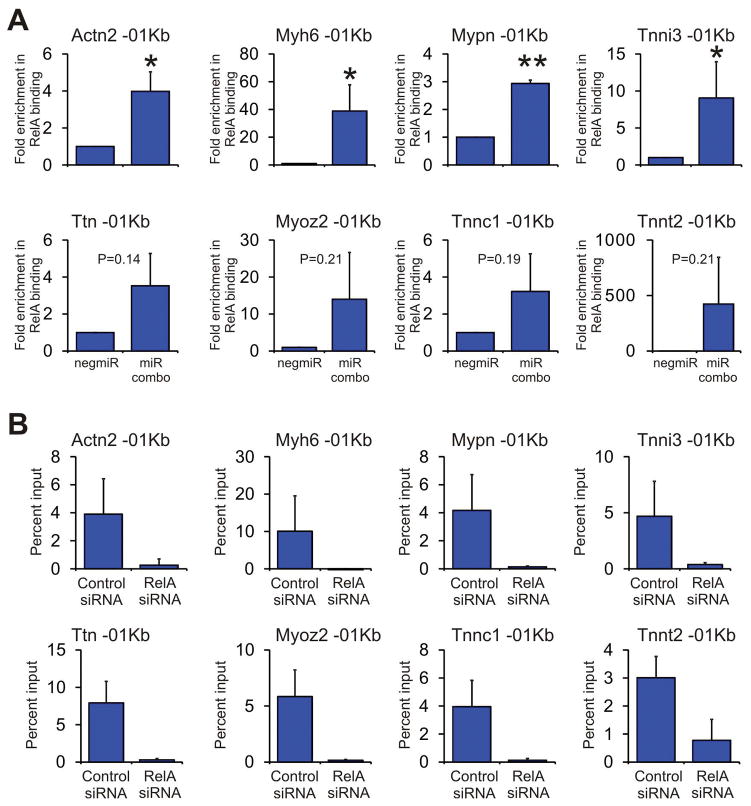
Figure 5. The NFκB subunit RelA binds to the promoters of cardiomyocyte maturation genes.
(A) Neonatal cardiac fibroblasts were transfected with negmiR or miR combo. After 7 days, chromatin DNA was subjected to ChIP analysis. Primers were designed to target the first 1Kb of the indicated cardiomyocyte sarcomere genes (represented by -01Kb). Results are presented as the fold enrichment in RelA binding where percent input of the negmiR control was taken to be 1. N=3 independent experiments. Data represented as Mean ± SEM. Comparisons are made between miR combo and negative control miR (negmiR), **P<0.01, *P<0.05, ns: not significant.
(B) Neonatal cardiac fibroblasts were transfected with miR combo and either a control siRNA or a siRNA that targeted RelA. After 7 days, chromatin DNA was subjected to ChIP analysis. Primers were designed to target the first 1Kb of the indicated cardiomyocyte sarcomere genes (represented by -01Kb). Results are presented as the percentage of chromatin input. N=3 independent experiments. Data represented as Mean ± SEM.