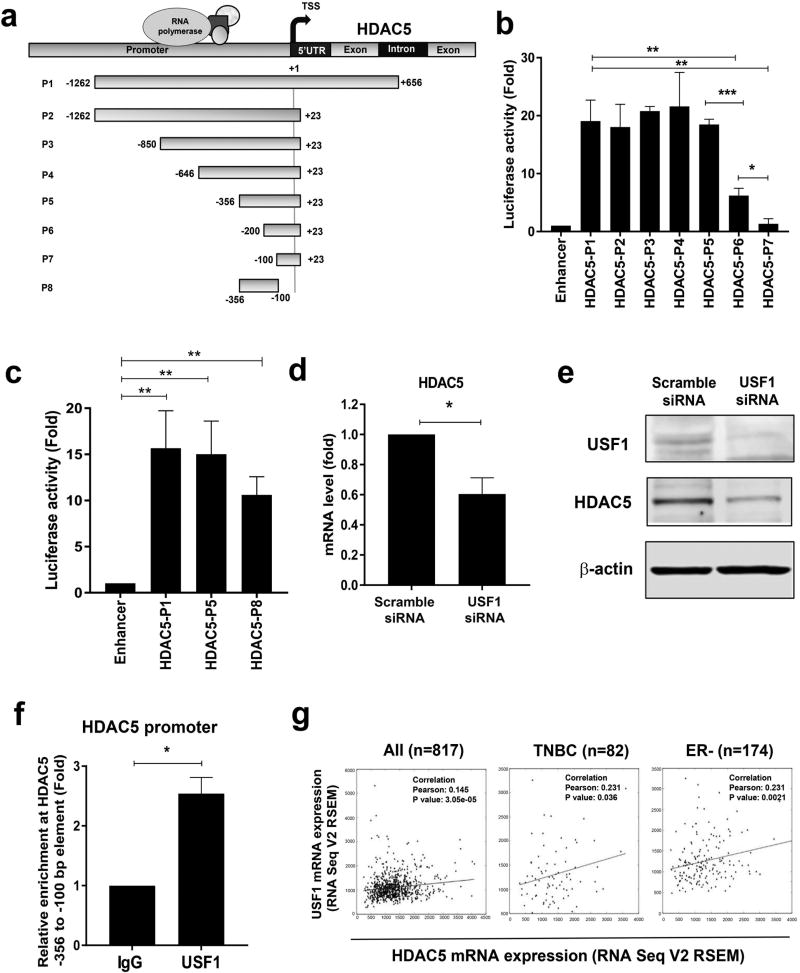
Figure 1. Analysis of transcriptional activity at HDAC5 promoter.
(a) Map of deletion constructs of HDAC5 promoter and coding region. TSS, transcription start site. (b) MDA-MB-231 cells were co-transfected with pGL2-Enhancer or pGL2-Enhancer-HDAC5 promoter elements and pRL-TK. Reporter gene activities were measured 48 h post-transfection. The relative luciferase activity of fragments P2-P7 was compared with that of full length P1. Transfection of pGL2-Enhancer plasmids was used as negative control. (c) MDA-MB-231 cells were co-transfected with pGL2-Enhancer or constructs of pGL2-Enhancer-HDAC5 promoter elements P1, P5 or P8 and pRL-TK. Reporter gene activities were measured 48 h post-transfection. Transfection of pGL2-enhancer was used as negative control. (d) MDA-MB-231 cells were transiently transfected with scramble or USF1 siRNA for 48 h. Effect of USF1 knockdown on HDAC5 mRNA expression was measured by quantitative PCR with β-actin as an internal control. (e) MDA-MB-231 cells were transfected with scramble or USF1 siRNA for 48 hr. Effect of USF1 knockdown on HDAC5 protein expression was examined by immunoblots with β-actin as loading control. (f) Quantitative ChIP analysis was used to determine the occupancy of USF1 protein at −356 to −100 bp element of HDAC5 promoter. (g) The Pearson correlation between mRNA expression of USF1 (y-axis) and HDAC5 (x-axis) in clinical TNBC or ER negative breast cancer specimens. Bars represent the mean of three independent experiments ± s.d. p<0.05 *, p<0.01 **, p<0.001 ***, Student’s t-test.