Figure 1. ADAM13 processing in neural crest cells.
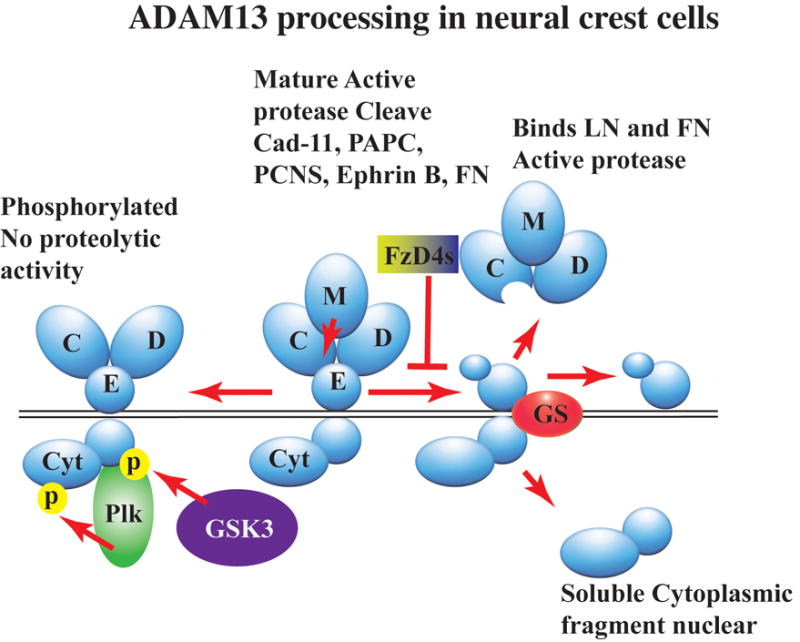
ADAMs are proteins that contain a Disintegrin (D) and Metalloprotease (M) domain. They are synthesized as pro-enzymes that get cleaved by Furin during the transit to the cell surface (elimination of the pro-domain not presented here). At the surface, ADAM13 cleaves both transmembrane proteins such as Cadherins, Protocadherins and Ephrin-B as well as extracellular matrix proteins such as fibronectin (FN). ADAM13 undergoes additional processing steps including self-proteolysis (right) within the cysteine (C) rich domain that releases the metalloprotease (M) domain associated with the disintegrin (D) and part of the cysteine (C) rich domain in the extracellular compartment. This protein remains proteolytically active and can bind to FN and Laminin (LN). The EGF repeat (E) remains associated with the transmembrane and cytoplasmic (Cyt) domains and becomes a substrate for γ-secretase (GS), which can cleave the cytoplasmic domain to allow it to translocate to the nucleus. Alternatively (left), the metalloprotease (M) domain can be cleaved to produce a non-proteolytic ADAM, which is the substrate of two kinases, GSK3 and Polo like kinase (Plk). The function of that form of ADAM13 is unknown, but the phosphorylation is critical for CNC migration.
