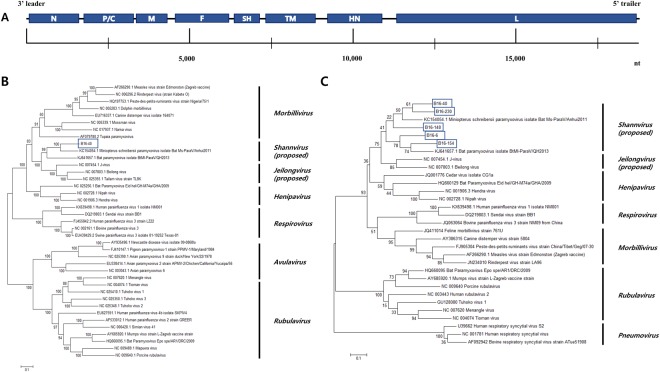
Figure 2.
(A) Genome organization of the bat paramyxovirus B16-40 isolated in this study. The 3′ leader and 5′ trailer regions are lined, 8 ORF regions for the viral proteins are indicated by blue-filled boxes. N, nucleocapsid protein; P, phosphoprotein; M, matrix protein; F, fusion protein; SH, small hydrophobic protein; TM, transmembrane protein; HN, attachment haemagglutinin neuraminidase glycoprotein; L, large protein. (B) Phylogenetic analysis based on the genomic nucleotide sequence of the bat paramyxovirus B16-40. Reference viruses belonging to Paramyxoviridae were included and aligned by MAFFT. The phylogenetic tree was generated by the maximum-likelihood method with 1,000 replicates of bootstrap sampling and the Kimura 2-parameter model using MEGA 6. Bat paramyxovirus B16-40 isolated in this study is indicated with the blue box. (C) Phylogenetic analysis based on partial RNA-dependent RNA polymerase (RdRp) nucleotide sequences of bat paramyxoviruses detected in the Korean bats and other reference viruses belonging to Paramyxoviridae. The phylogenetic tree was generated by the maximum-likelihood method with 1,000 replicates of bootstrap sampling and the Kimura 2-parameter model using MEGA 6. Korean bat paramyxoviruses are indicated by blue boxes.