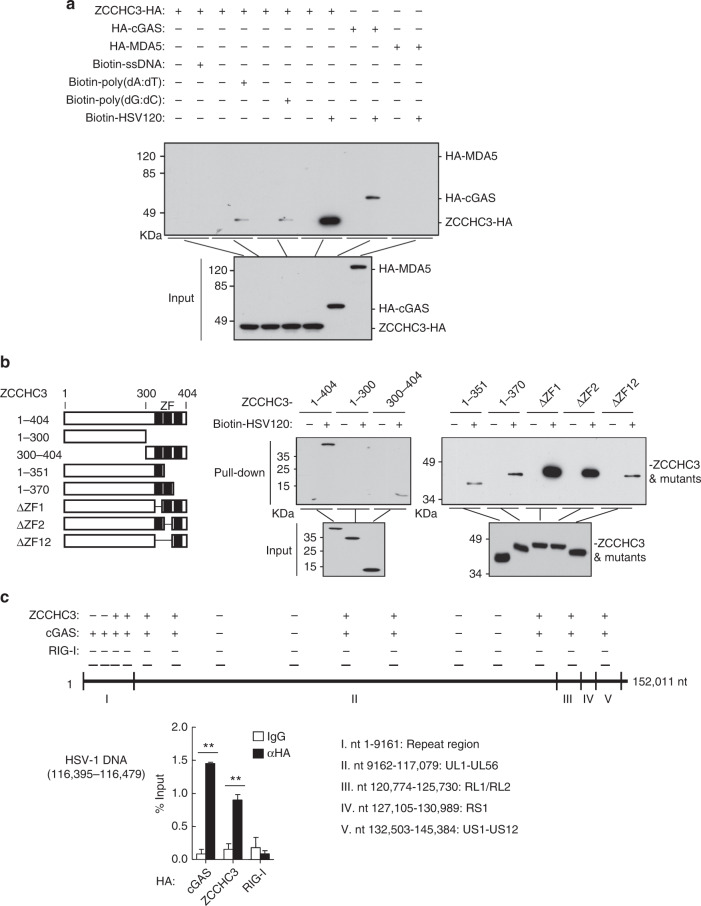
Fig. 5.
ZCCHC3 binds to dsDNA. a ZCCHC3 binds to dsDNA. HEK293 cells were transfected with the indicated plasmids. Twenty hours later, the cell lysates were incubated with the indicated biotinylated nucleic acids and streptavidin-Sepharose beads for in vitro pull-down assays. The bound proteins were then analyzed by immunoblots with anti-HA. b ZCCHC3 binds to dsDNA through its C-terminal ZF domains. HEK293 cells were transfected with the indicated plasmids. Twenty hours after transfection, the cell lysates were incubated with biotinylated-HSV120 and streptavidin-Sepharose beads. The bound proteins were analyzed by immunoblots with anti-Flag. A schematic representation of ZCCHC3 and its truncation mutants was shown on the left. c ZCCHC3 and cGAS but not RIG-I bind to HSV-1 DNA of infected cells. HEK293 cells were transfected with HA-tagged ZCCHC3, cGAS, and RIG-I. Twenty hours after transfection, cells were infected with HSV-1for 3 h. Cell lysates were then immunoprecipitated with control IgG or anti-HA. The protein-bound DNAs were extracted and analyzed by qPCR analysis with primers corresponding to the indicated regions of HSV-1 genome. Positive ( + ) and negative (-) detections were shown at the top of the schematic presentation of the HSV-1 genome. A representative qPCR results were shown at the left. **P < 0.01 (unpaired t test). Data are representative of three experiments with similar results