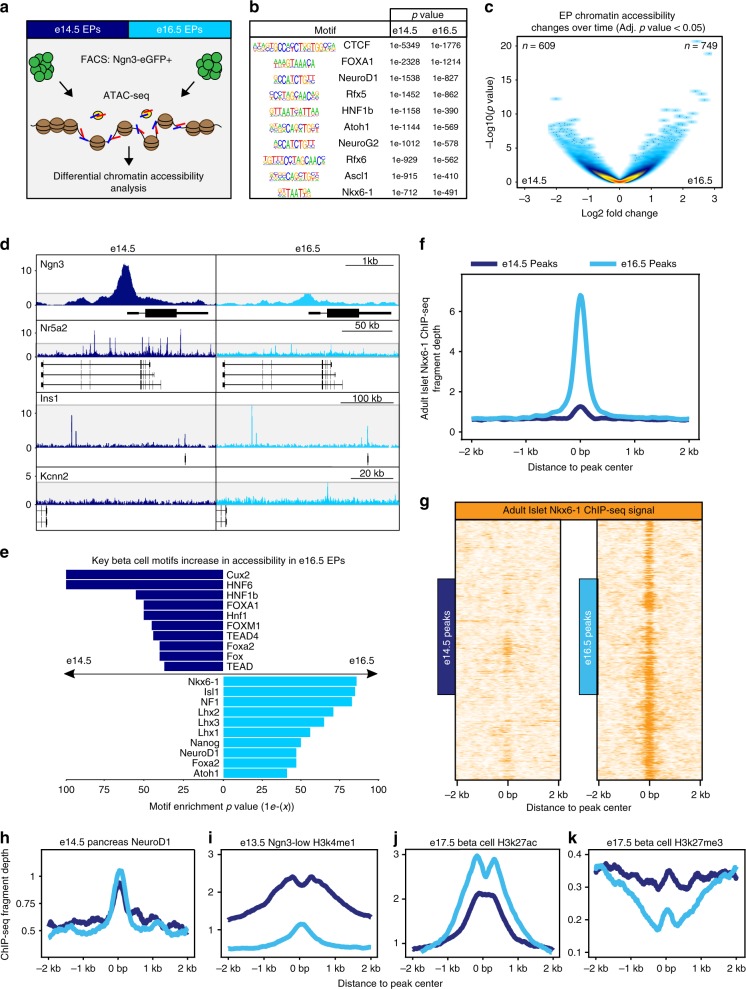
Fig. 5.
Shifts in chromatin accessibility promote the potential of endocrine progenitors for the beta cell fate at later stages of embryonic development. a Scheme of ATAC-seq experiments. Pancreata from Ngn3-eGFP mice were dissected at e14.5 or e16.5 and Ngn3-eGFP + cells were isolated using FACS for ATAC-seq. n = 3 biological replicates. b Motifs globally enriched in accessible chromatin regions of e14.5 and e16.5 EPs. c The chromatin landscape changes from e14.5 to e16.5. Volcano plot showing chromatin accessibility. Adjusted p-value < 0.05. d ATAC-seq tracks highlighting the locus of Ngn3, Nr5a2, Ins1, and Kcnn2 from e14.5 (shown in navy) and e16.5 (in light blue) EPs, after normalization and combination of all biological replicates. Genomic loci: Ngn3, chr10:61,593,975-61,597,673. Nr5a2, chr1:138,737,109-138,913,736. Ins1, chr19:52,066,906-52,389,227. Kcnn2, chr18:45,839,320-45,911,754. e Top differentially enriched motifs in accessible chromatin regions of e14.5 (in navy) and e16.5 EPs (in light blue). f Adult islet Nkx6-1 ChIP-seq fragment coverage histogram (per base pair per peak) counted across the e14.5 and e16.5 EP differentially accessible peaks from Fig. 5c. Data from GSM1006208. g Adult islet Nkx6-1 ChIP-seq signal (reads) heatmap across the e14.5 and e16.5 EP differentially accessible peaks, as show in Fig. 5f. Rows are peaks, and each peak is centered and signal visualized across a 4 kb window. Low ChIP-seq signal is colored as white, and high signal (reads) is colored orange. h The e14.5 whole pancreas NeuroD1 ChIP-seq (GSM2532979) fragment coverage histogram (per base pair per peak) counted across the e14.5- and e16.5-specific EP peaks. i The e13.5 Ngn3-GFP low H3K4me1 ChIP-seq fragment coverage histogram (per base pair per peak) counted across the e14.5- and e16.5-specific EP peaks. ChIP-seq data from GSE84324. j The e17.5 beta cell H3K27ac ChIP-seq fragment coverage histogram (per base pair per peak) counted across the e14.5- and e16.5-specific EP peaks. ChIP-seq data from GSE84324. k The e17.5 beta cell H3K27me3 ChIP-seq fragment coverage histogram (per base pair per peak) counted across the e14.5- and e16.5-specific EP peaks. ChIP-seq data from GSE84324. See also Supplementary Figure 15