Abstract
Autologous transplantation of hematopoietic stem cells transduced with a lentiviral vector (LV) expressing an anti-sickling HBB variant is a potential treatment for sickle cell disease (SCD). With a clinical trial as our ultimate goal, we generated LV constructs containing an anti-sickling HBB transgene (HBBAS3), a minimal HBB promoter, and different combinations of DNase I hypersensitive sites (HSs) from the locus control region (LCR). Hematopoietic stem progenitor cells (HSPCs) from SCD patients were transduced with LVs containing either HS2 and HS3 (β-AS3) or HS2, HS3, and HS4 (β-AS3 HS4). The inclusion of the HS4 element drastically reduced vector titer and infectivity in HSPCs, with negligible improvement of transgene expression. Conversely, the LV containing only HS2 and HS3 was able to efficiently transduce SCD bone marrow and Plerixafor-mobilized HSPCs, with anti-sickling HBB representing up to ∼60% of the total HBB-like chains. The expression of the anti-sickling HBB and the reduced incorporation of the βS-chain in hemoglobin tetramers allowed up to 50% reduction in the frequency of RBC sickling under hypoxic conditions. Together, these results demonstrate the ability of a high-titer LV to express elevated levels of a potent anti-sickling HBB transgene ameliorating the SCD cell phenotype.
Keywords: sickle cell disease, lentiviral vectors, gene therapy
Introduction
Sickle cell disease (SCD) is a severe genetic disorder affecting ∼312,000 newborns worldwide annually.1 A single point mutation in the adult β-globin (HBB) gene causes a Glu > Val amino acid substitution in the β-globin chain (βS-globin). The sickle hemoglobin (HbS, α2βS2) has the propensity to polymerize under deoxygenated conditions, resulting in the production of sickle-shaped red blood cells (RBCs) that can cause occlusions of blood vessels, leading to impaired oxygen delivery to tissues, respiratory complications, organ damage, and early mortality. Additionally, sickled RBCs are prone to hemolysis; thus, the clinical phenotype is also characterized by hemolytic anemia. Current treatments include life-long RBC transfusions and hydroxyurea, a drug that increases fetal Hb (HbF, α2γ2) synthesis.2 Indeed, the clinical course of SCD is improved when fetal HBG genes are highly expressed, as seen in patients with naturally occurring mutations leading to hereditary persistence of fetal hemoglobin (HPFH). In SCD, γ-globin exerts a potent anti-sickling function by competing with the sickle βS-globin for incorporation in Hb tetramers and by inhibiting HbS polymerization. However, pharmacological treatments increasing HbF levels are not equally effective in all patients.2
The only definitive cure for SCD patients is allogenic hematopoietic stem cell (HSC) transplantation. However, HSC transplantation from an HLA-matched related donor is available only to a fraction of patients.3 Transplantation of HSCs from matched unrelated donors are associated with a higher risk of graft-versus-host-disease, transplant rejection and infections.3 With the advent of HBB expressing lentiviral vectors (LVs), transplantation of genetically modified autologous HSCs holds promise of circumventing the need for suitable donors and the morbidity and mortality associated with allogenic transplantation.
LV-based ex vivo gene therapy strategies require the stable transfer of an anti-sickling HBB globin transgene in the patient’s long-term repopulating HSCs and high, sustained, and regulated expression of the therapeutic globin chain in their erythroid progeny. Several LVs have been developed and tested in murine models of SCD and patient hematopoietic stem progenitor cells (HSPCs).4, 5, 6 In these vectors, an anti-sickling HBB transgene (HBG or βT87Q and βAS3 HBB anti-sickling variants) is placed under the transcriptional control of the HBB promoter and key regulatory elements from the 16-kb human β-locus control region (βLCR), which is essential for high and regulated expression of the endogenous HBB gene family.7 Since LVs cannot accommodate the entire LCR, only the three most transcriptionally potent out of the five DNase I hypersensitive sites (HS2, HS3, and HS4) were selected and reduced in size to fit into the vector packaging capacity. The combination of minimal core elements of HS2, HS3, and HS4 (each of them 0.2 to 0.4 kb long) was associated with low transgene expression levels, positional variegation, and transcriptional silencing, whereas extended HSs sustained high HBB-like globin expression.8, 9 Sequences flanking the HS core elements might indeed be required for synergistic activation of HBB-like globin gene expression.10 Therefore, β-globin expressing LVs are exceptionally large, harboring a 2.6 to 3.4 kb “mini-LCR” containing extended HS2, HS3, and HS4.4, 5, 6 The large size of these vectors makes their production challenging and might affect gene transfer efficiency, particularly in HSPCs.8
There have been three clinical trials for SCD using LVs expressing an anti-sickling HBB-like globin gene (see Cavazzana et al.11 and Ferrari et al.12). The first SCD patient to have undergone gene therapy showed a good correction of the clinical phenotype and the biological hallmarks of SCD.13 However, in a parallel study using the same LV, gene marking in the peripheral blood was low in all treated SCD subjects, with no evidence of clinical benefit (J. Kanter et al., 2011, Am. Soc. Hem., conference). Therefore, the development of high-titer LVs able to transduce a good proportion of HSCs and drive high levels of HBB-like expression is still a critical issue to achieve therapeutic efficacy in SCD patients. In particular, the choice of βLCR regulatory elements able to drive high levels of HBB-like expression without compromising the vector titer is critical.
Here, we show that a compact high-titer LV containing only the HS2 and HS3 βLCR elements is highly efficient in transducing a large proportion of SCD HSPCs derived from bone marrow or mobilized in the peripheral blood of SCD patients and expressed therapeutic levels of a potent anti-sickling HBB transgene.
Results
Design and Characterization of LVs Expressing an Anti-sickling Human HBB Transgene
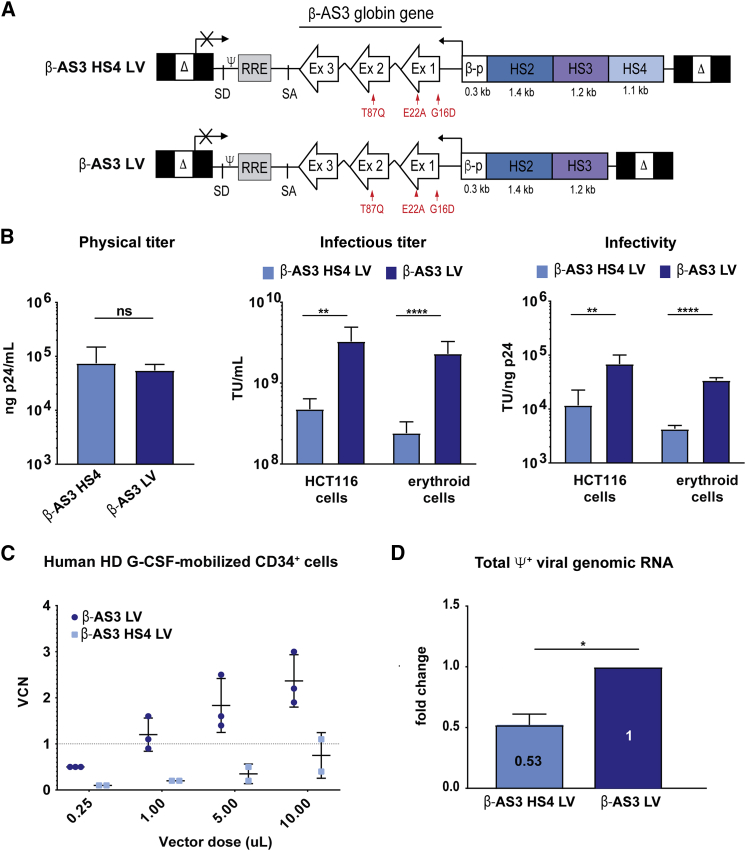
We generated two LVs carrying an anti-sickling human HBB transgene (HBBAS3) under the transcriptional control of a minimal HBB promoter and either two or three HSs from the human βLCR: HS2 and HS3 (β-AS3 LV) and HS2, HS3, and HS4 (β-AS3 HS4 LV) (Figure 1A). The HBBAS3 gene contains three mutations14 causing three potentially beneficial “anti-sickling” amino-acidic substitutions (G16D, E22A, T87Q) in the LV-derived HBB chain (βAS3): A22 and Q87 impair, respectively, the axial and lateral contacts necessary for the formation of HbS polymers, and D16 increases the affinity to HBA chains, thus conferring to βAS3 a competitive advantage for the incorporation in the Hb tetramers (Figure 1A).14
Figure 1.
Characterization of β-AS3 HBB-Expressing LVs
(A) Schematic representation of β-AS3 HS4 and β-AS3 lentiviral vectors. Δ, deleted HIV-1 U3 region; SD and SA, HIV splicing donor and acceptor sites; ψ, HIV-1 packaging signal; RRE, HIV-1 Rev responsive element; Ex, exons of the human HBB; βp, promoter of HBB; HS2, 3, and 4, DNase I hypersensitive site 2, 3, and 4 of human HBB LCR; red arrows indicate the mutations introduced in exon 1 (generating amino acid substitutions G16D and E22A) and exon 2 (generating amino acid substitution T87Q). (B) The histograms show the physical and infectious titers and infectivity of β-AS3 HS4 and β-AS3 LVs. Infectious titer and infectivity were measured in HTC116 (five different preparations for each vector) and K562 and HEL erythroid cell lines (two viral preparations per vector). (C) Vector copy number (VCN) in G-CSF-mobilized CD34+ cells from healthy donors (HDs). HSPCs were transduced with increasing amounts of three and two preparations of β-AS3 and β-AS3 HS4 LVs, respectively. Cells were grown in liquid culture, and after 1 week, VCN was determined. A linear correlation between β-AS3 vector dose and VCN is obtained, whereas a modest increase in VCN was achieved by transducing HSPCs with higher amounts of β-AS3 HS4 vector preparations. (D) Relative quantification of the ψ+ viral genomic RNA produced by HEK293T packaging cells (n = 3) by qRT-PCR. The β-AS3 sample was used as a calibrator. Data were plotted as mean ± SD (unpaired t test). *p ≤ 0.05, **p ≤ 0.01, ****p ≤ 0.0001, ns, not significant.
Virus glycoprotein G (VSV-G) pseudotyped, third generation LVs, were produced by standard transient transfection of HEK293T cells and concentrated by ultra-centrifugation. The physical titer of the two vectors, determined by measuring the viral p24 protein, was similar, indicating that a comparable amount of total viral particles was present in the β-AS3 LV and β-AS3 HS4 vector preparations (Figure 1B). We then transduced HCT116 cells and human erythroid (HEL and K562) cell lines to determine the infectious titer by measuring the average number of vector copies integrated per genome (vector copy number; VCN). The infectious titer of β-AS3 LV was ∼8-fold higher, compared to β-AS3 HS4 LV (Figure 1B). As a consequence, β-AS3 LV showed a markedly increased infectivity, measured as ratio between the infectious and the physical titer (Figure 1B). We then transduced human granulocyte colony-stimulating factor (G-CSF)-mobilized peripheral blood (mPB) HSPCs from healthy donors (HDs) with increasing amounts of vector preparations. Notably, β-AS3 LV showed a superior gene transfer efficiency compared to β-AS3 HS4, leading to up to 2.40 ± 0.58 vector copies per cell at the highest vector dose (Figure 1C).
Given these results, we next investigated the potential detrimental effects of the HS4 element on viral titer and infectivity. Total RNA was extracted from HEK293T packaging cells transfected with β-AS3 and β-AS3 HS4 transfer vectors and LV packaging plasmids. qRT-PCR analysis revealed that the total viral genomic RNA containing the ψ+ packaging signal was significantly higher for β-AS3 LV compared to the HS4-containing LV (Figure 1D). In addition, by using 3′ rapid amplification of cDNA ends (RACEs)-PCR followed by Sanger sequencing, we detected a truncated β-AS3 HS4 viral genomic transcript (Figure S1A). We identified a canonical poly-adenylation signal (AATAAA) in the HS4 element, likely responsible of the premature truncation of the β-AS3 HS4 viral transcripts (Figure S1A). However, qRT-PCR showed a comparable fraction of full-length viral RNA (∼20% of the total ψ+ viral genomic RNA) for both β-AS3 and β-AS3 HS4 LVs (Figure S1B).
To understand the reasons for the significant drop in titer cause by the 1.1-kb-long HS4, we constructed the β-AS3 HS4 core LV harboring a short 304-bp HS4 fragment containing the core HS4 element10 and lacking the polyadenylation site mapped in the long HS4 element (Figure S1C). Although the shortening of the HS4 element restored most of the total ψ+ viral genomic RNA levels, it did not increase significantly the infectious titer or infectivity in HCT116 cells, thus indicating that even the core HS4 elements contain sequences that are detrimental to the vector performance (S1D and S1E).
β-AS3 Transduction Leads to Highly Efficient Gene Transfer in SCD Bone Marrow HSPC
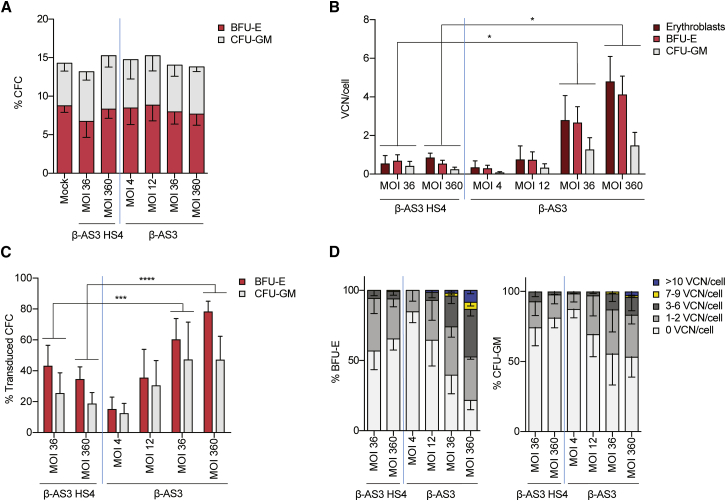
In order to evaluate gene transfer efficiency of the β-AS3 and β-AS3 HS4 LVs, we transduced SCD patient bone marrow (BM) HSPCs samples with an equal number of infecting particles (MOI 4–360; n = 2–6; two donors). HSPCs were either differentiated in liquid culture toward the erythroid lineage or plated in semi-solid medium containing cytokines supporting the clonal growth and differentiation of erythroid (burst forming unit-erythroid [BFU-E]) and colony forming unit-granulocyte-monocyte (CFU-GM) progenitors (colony-forming cell [CFC] assay). Lentiviral transduction did not affect erythroid proliferation and differentiation or clonogenic potential of transduced HSPCs (Figures 2A and S2 and data not shown).
Figure 2.
Gene Transfer Efficiency in SCD Patient-Derived BM HSPCs Transduced with β-AS3 HS4 and β-AS3 LVs
(A) Percentages of CD34+ HSPCs that gave rise to BFU-E and CFU-GM in CFC assay. Values shown are mean ± SEM of three to five independent experiments. (B) Average VCN/cell in bulk populations of erythroblasts grown in liquid culture, and pools of BFU-E and CFU-GM derived from β-AS3 HS4- and β-AS3-transduced SCD BM HSPCs. Values shown are mean ± SEM of two to six independent experiments (n = 2 donors). *p ≤ 0.05 (unpaired t test). (C) Percentage of vector+ BFU-E and CFU-GM derived from SCD BM HSPCs transduced with β-AS3 HS4. Values shown are mean ± SEM of three to six independent experiments (n = 2 donors). ***p ≤ 0.001, ****p ≤ 0.0001 (chi-squared test). (D) Clonal analysis of VCN in BFU-E and CFU-GM colonies derived from SCD BM HSPCs transduced with β-AS3 HS4 or β-AS3. Values shown are mean ± SEM of three to six independent experiments (n = 2 donors). A higher proportion of colonies harboring >3 VCN was observed for β-AS3 LV at MOIs of 36 and 360 (p ≤ 0.001; chi-squared test).
Negligible gene transfer efficiency was observed using β-AS3 HS4 with an MOI lower than 36 (data not shown). HSPC transduction with β-AS3 HS4 at an MOI of 36 led to the integration of 0.55 ± 0.41, 0.70 ± 0.31, and 0.43 ± 0.23 vector copies per cell in erythroblasts, and pools of BFU-E and CFU-GM, respectively. A 10-fold increase in β-AS3 HS4-infecting particles did not increase the gene transfer efficiency (Figure 2B). In contrast, β-AS3 was able to transduce HSPCs even at low MOIs (4 and 12; Figure 2B). The VCN of β-AS3 was positively correlated with the MOIs, rising up to 4.81 ± 1.29 in erythroblasts, 4.13 ± 0.95, in BFU-E and 1.49 ± 0.67 in CFU-GM at an MOI of 360. Notably, at the same MOIs β-AS3 transduction efficiency was significantly higher, as compared to β-AS3 HS4 (Figure 2B).
To assess precisely the frequency of transduced HSPCs, we measured the gene transfer efficiency in individual BFU-E and CFU-GM (n = 3–6; 2 donors). When HSPCs were transduced using the same vector doses, a higher proportion of transduced CFCs was observed for β-AS3 compared to β-AS3 HS4 (Figure 2C). We observed up to 82% and 67% of β-AS3-transduced BFU-E and CFU-GM, respectively. In contrast, β-AS3 HS4 did not transduce >∼40% of CFCs (Figure 2C). Samples containing <40% of transduced CFCs (e.g., β-AS3 HS4, MOI 360 and 36; and β-AS3, MOI 4 and 12) displayed a similar VCN distribution, with most of the transduced BFU-E and CFU-GM harboring 1 to 2 vector copies per genome. Notably, the high frequency of transduction reached using β-AS3 at the highest doses (e.g., MOI 36 and 360) was accompanied with a significantly higher proportion of progenitors harboring >3 vector copies per cell (from 10% to 65%), compared to β-AS3 HS4 (<5%) (Figure 2D).
β-AS3 Outperforms β-AS3 HS4 in the Transduction of Long-Term Repopulating HSCs
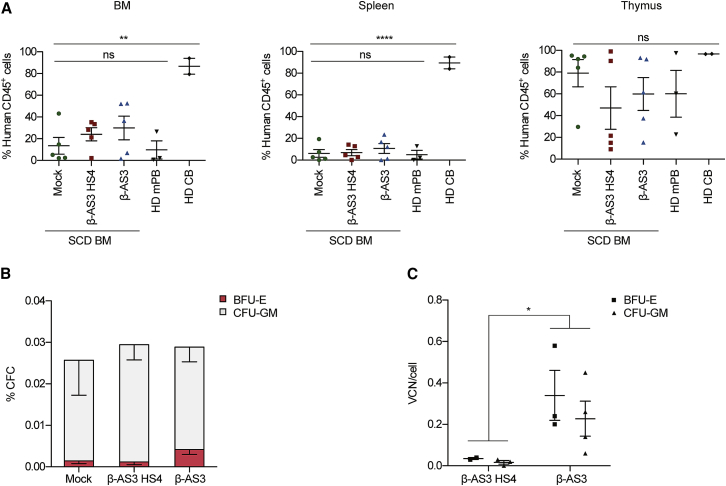
We then compared gene transfer efficiency of the β-AS3 HS4 and β-AS3 LVs in long-term repopulating SCD HSCs. BM-derived HSPCs from an SCD patient were transduced at an MOI of 360 and injected into NOD scid gamma (NSG) immunodeficient mice after busulfan-based conditioning (n = 5 mice per vector). As expected, the transduction efficiency of the input SCD HSPC populations was higher for β-AS3 than for β-AS3 HS4, as assessed in vitro in BFU-E and CFU-GM pools (0.26 and 0.18 β-AS3 HS4 vector copies in BFU-E and CFU-GM; 2.03 and 0.66 β-AS3 vector copies in BFU-E and CFU-GM). As controls, we transplanted mock-transduced SCD HSPCs (n = 5 mice) and adult G-CSF-mPB or cord blood (CB) HSPCs obtained from HDs (n = 5 mice).
Mice were euthanized and hematopoietic organs were analyzed for the engraftment of human cells, calculated as percentage of human CD45+ cells in the total human and murine CD45+ cell populations. The engraftment was similar among mock and transduced conditions in BM, spleen, and thymus (Figure 3A). Comparable frequencies were observed upon transplantation of HD HSPCs, except for a higher engraftment of human CD45+ cells in the spleen and thymus of mice transplanted with CB-derived HSPCs. We detected a similar distribution of B, T, and myeloid cells in the CD45+ cell populations across the different conditions (Figure S3).
Figure 3.
β-AS3 HS4 and β-AS3 Transduction Efficiencies in Long-Term Repopulating SCD HSCs
(A) Engraftment of human cells in NSG mice transplanted with mock- and LV-transduced SCD BM CD34+ cells (MOI = 360, n = 5 mice for each group) and HD mPB (n = 3) and HD CB (n = 2) HSPCs 15 weeks after transplantation. Engraftment is represented as percentage of human CD45+ cells in the total murine and human CD45+ cell population, in BM, spleen, and thymus. Values shown are mean ± SEM; ns, not significant (one-way ANOVA test). (B) Human hematopoietic progenitor content in BM mononuclear cells (MNC) derived from mice transplanted with either mock- or β-AS3 HS4- and β-AS3-transduced SCD BM HSPCs. We plotted the percentage of BM MNC giving rise to BFU-E and CFU-GM. Values shown are mean ± SEM of four samples per condition. (C) VCN/cell in pools of BFU-E- and CFU-GM-derived from total BM MNC, obtained from mice injected with SCD BM HSPCs transduced with β-AS3 HS4 (n = 2 and n = 3 for BFU-E and CFU-GM, respectively) and β-AS3 (n = 3 and n = 4 for BFU-E and CFU-GM, respectively) LVs at an MOI of 360. Values shown are mean ± SEM. *p ≤ 0.05; **p < 0.01; ****p < 0.0001 (unpaired t test).
Total BM cells were isolated and subjected to a CFC assay to assess the gene transfer efficiency in HSC-derived BFU-E and CFU-GM pools. Clonogenic potential was similar between mock and transduced samples (Figure 3B). The average β-AS3 HS4 VCN per cell was 0.04 ± 0.001 in BFU-E and 0.02 ± 0.001 in CFU-GM, whereas the β-AS3-transduced CFCs displayed a significantly higher VCN/cell (0.34 ± 0.12 in BFU-E and of 0.23 ± 0.08 in CFU-GM; Figure 3C).
Taken together, these results confirmed the superior gene transfer efficiency of the β-AS3 vector in long-term repopulating HSCs compared to the β-AS3 HS4 LV harboring an additional βLCR HS site.
β-AS3 LV-Derived Transgene Expression Ameliorates the SCD Cell Phenotype In Vitro
To compare the therapeutic potential of the β-AS3 and β-AS3 HS4 LVs in terms of transgene expression and correction of the SCD phenotype, we differentiated in vitro-transduced SCD BM HSPCs into mature RBCs (n = 3–4; two donors). Erythroid differentiation was monitored over time by flow cytometry and morphological analyses. Downregulation of early erythroid markers CD36 and CD71 and upregulation of the late erythroid marker glycophorin A (GYPA) occurred similarly in mock and transduced samples (Figures S2A and S2B). RBC enucleation was obtained at day 20-post differentiation for all the samples, as demonstrated by the loss of cells staining with the nuclear dye DRAQ5 and by morphological analyses (Figures S2A–S2C).
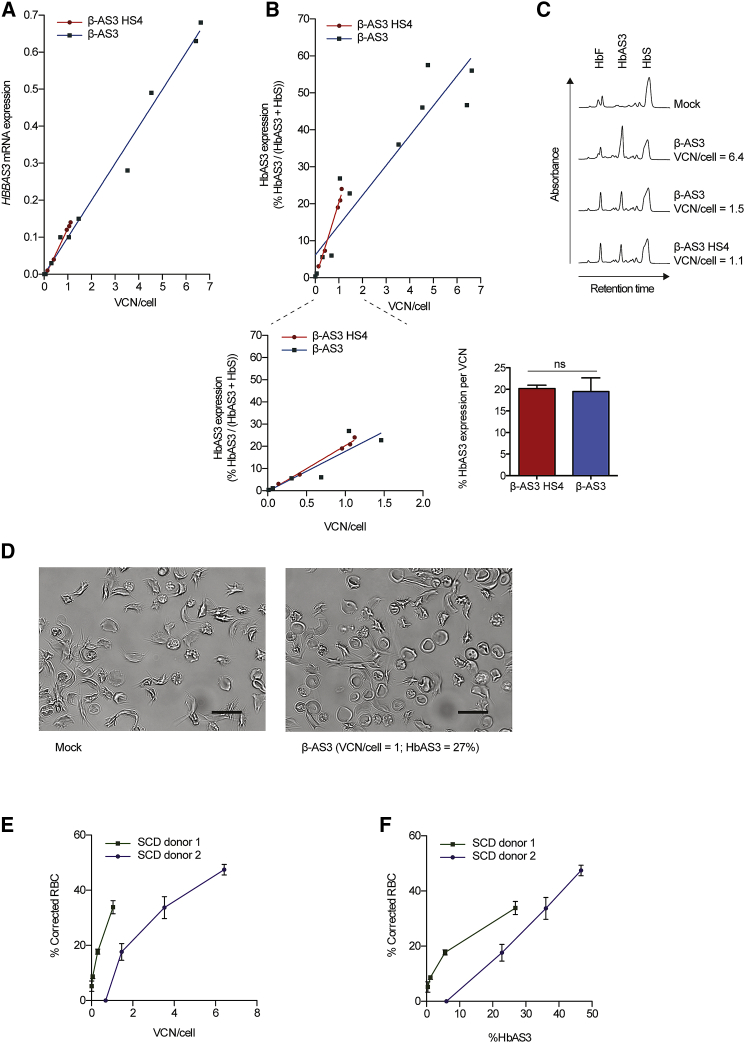
HBBAS3 transgene mRNA expression was measured by qRT-PCR in erythroblasts derived from β-AS3- and β-AS3 HS4-transduced HSPCs. We observed a strong correlation between transgene expression and average VCN for both β-AS3 HS4 and β-AS3 LVs (Pearson correlation, R = 0.99, p < 0.0001; Figure 4A). Despite the lack of the HS4 element in the β-AS3 construct, no statistically significant differences in HBBAS3 expression were observed between the two vectors in erythroblasts derived from HSPCs harboring a similar VCN (Figure 4A). Given the higher transduction efficiency, β-AS3 LV-derived HBBAS3 expression was overall higher in erythroblasts derived from SCD HSPCs transduced at high MOIs (e.g., 36 and 360), as compared to β-AS3 HS4-transduced samples (β-AS3 mRNA levels, 0.37 ± 0.11 and 0.09 ± 0.03, respectively). Similar results were obtained in BFU-E transduced with β-AS3 HS4 and β-AS3 LVs (Figure S4A).
Figure 4.
Transgene Expression in Erythroid Cells Derived from β-AS3 HS4- and β-AS3-Transduced SCD BM HSPCs and Phenotypic Correction of SCD RBCs by β-AS3
(A) HBBAS3 mRNA expression was measured by RT-qPCR in erythroblasts at day 13 post-induction of erythroid differentiation, normalized using the endogenous βS-globin gene (HBBS), and correlated with VCN/cell. The slopes of the linear regression lines for samples derived from β-AS3 HS4- and β-AS3-transduced HSPCs were not significantly different (p = 0.2922). Equations that define the best fit lines were as follows: y = 0.13x − 0.011 (R2 = 1.0 and p < 0.0001) for β-AS3 HS4 samples and y = 0.10x − 0.00059 (R2 = 0.99 and p < 0.0001) for β-AS3 samples. 5 β-AS3 HS4- and 10 β-AS3-transduced samples were analyzed (n = 2 donors). (B) HbAS3 was quantified by cation-exchange HPLC in RBCs at day 20 of erythroid differentiation. The percentage of HbAS3 in the total adult Hb (HbAS3 + HbS) was correlated with the average VCN/cell. Top, the slopes of the linear regression lines for RBC samples derived from β-AS3 HS4- and β-AS3-transduced HSPCs were not significantly different (p = 0.1446) (line-of-best-fit equation, y = 20.79x − 0.49, R2 = 0.9905 and p = 0.0004 for β-AS3 HS4 samples and y = 8.08x + 6.10, R2 = 0.8776 and p < 0.0001 for β-AS3 samples). 5 β-AS3 HS4- and 11 β-AS3-transduced samples were analyzed (n = 2 donors). Bottom right, plot of HbAS3 expression in RBCs derived from β-AS3 HS4- and β-AS3-transduced samples harboring a similar VCN/cell. Bottom left, HbAS3 expression per VCN/cell. Data were plotted as mean ± SEM (unpaired t test). ns, not significant. (C) HPLC chromatograms of RBC lysates obtained after 20 days of erythroid differentiation of mock- and LV-transduced SCD BM HSPCs. (D–F) In vitro sickling assay measuring the proportion of sickled RBC under hypoxic conditions (0% O2). Representative photomicrographs of RBCs derived from mock- and β-AS3-transduced samples at 0% of O2 are shown (D); SCD donor 1. Scale bars, 20 μm. The percentage of corrected RBCs was correlated to the VCN/cell (E) or to HbAS3 expression (F) for two SCD donors. The proportion of corrected cells was calculated as the percentage of non-sickled cells in the transduced samples, minus the percentage of non-sickled cells in the corresponding mock-transduced sample. Values are mean ± SEM of three and four microscopic fields for donor 1 and donor 2, respectively. For donor 2, a higher VCN and transgene expression were necessary to achieve a frequency of corrected RBCs comparable to the value observed for donor 1. This difference may be due to the coinheritance of different genetic disease modifiers along with the SCD mutation.
The expression of the Hb tetramer containing the therapeutic βAS3globin-chain (HbAS3) was detected in mature RBCs by HPLC and was accompanied by a reduction in the levels of HbS containing βS-chains (Figures 4B and 4C). HbAS3 expression was positively correlated with the VCN/cell of β-AS3 HS4 and β-AS3 LVs (Pearson correlation, R = 0.94, p < 0.0001 for β-AS3 and R = 1, p < 0.001 for β-AS3 HS4; Figure 4B). We observed no significant differences in HbAS3 content in RBCs derived from HSPCs harboring similar β-AS3 HS4 and β-AS3 vector copies (Figures 4B and 4C). Indeed, HbAS3 expression normalized per VCN was similar for the two vectors (Figure 4B). Notably, HSPC transduction with the β-AS3 LV led to greater protein expression at MOIs higher than 36 compared to β-AS3 HS4 (39.26% ± 6.92% for β-AS3 and 14.87% ± 4.09% for β-AS3 HS4; p < 0.05). Similarly, no differences in HbAS3 expression were observed in BFU-E harboring similar β-AS3 HS4 and β-AS3 VCNs (Figure S4B).
The high levels of β-AS3-derived anti-sickling transgene expression prompted us to assess the ability of β-AS3 LV to correct the SCD cell phenotype. To measure the frequency of sickling and corrected RBCs, we performed an in vitro sickling assay under hypoxic conditions. SCD BM HSPCs were transduced with increasing β-AS3 vector doses and differentiated in mature RBCs (n = 2 donors). Upon deoxygenation, the frequency of sickling RBCs was >80% in mock-transduced samples (Figure 4D). In marked contrast, HSPC transduction with the β-AS3 LV led to a decreased frequency of sickling RBCs and to significant proportions of biconcave RBCs under 0% O2 (Figure 4D). The percentage of corrected (i.e., non-sickling) RBCs increased gradually with the VCN, reaching 34% and 47% of corrected RBCs for donor 1 and 2, respectively (Figure 4E). Notably, the extent of the RBC correction was positively correlated with the HbAS3 production (Figure 4F).
β-AS3 LV Efficiently Transduces SCD Mobilized HSPCs
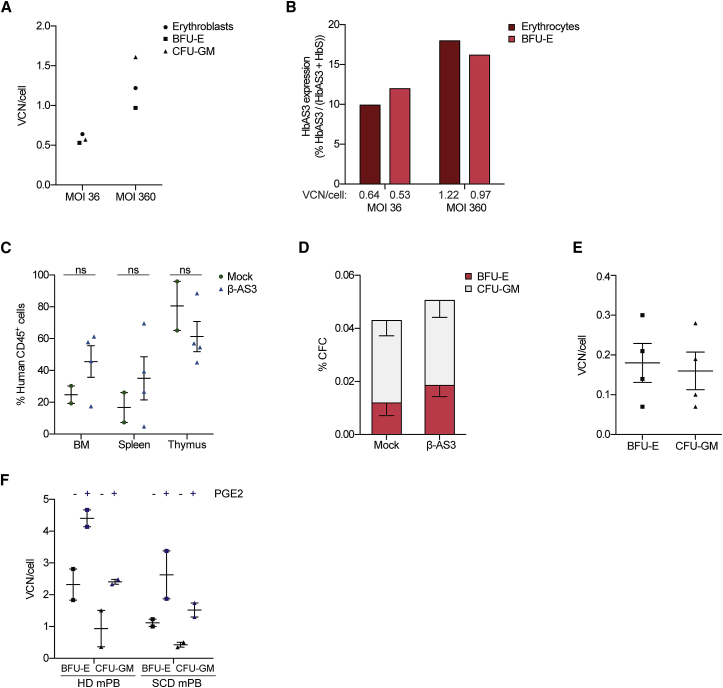
We recently showed that HSPCs can be safely and efficiently mobilized from SCD patients by using Plerixafor.15 To verify if mobilized SCD HSPCs are permissive to β-AS3 LV transduction, these cells were transduced at an MOI of 36 and of 360. The average β-AS3 VCN per cell increased with the vector dose, reaching 1.22 in erythroblasts, 0.97 in BFU-E and 1.61 in CFU-GM at the highest MOI (n = 1 donor; Figure 5A). Concomitantly, HbAS3 expression reached 17.97% of total Hb in in vitro generated mature RBCs and 16.18% in BFU-E at an MOI of 360 (Figure 5B).
Figure 5.
In Vitro and In Vivo Gene Transfer Efficiency in Cells Derived from SCD Plerixafor-Mobilized β-AS3-Transduced HSPCs
(A) VCN/cell in bulk populations of erythroblasts and in pools of BFU-E and CFU-GM derived from β-AS3-transduced SCD Plerixafor mobilized HSPCs. Transductions were performed using MOIs of 36 and 360. (B) HbAS3 expression was quantified by cation-exchange HPLC in BFU-E and RBCs. Average VCN/cell is indicated. (C) Engraftment of human CD45+ cells derived from SCD mobilized samples in BM, spleen, and thymus of NSG mice 18 weeks after transplantation. Plerixafor-mobilized SCD HSPCs were transduced with β-AS3 LV at an MOI of 100. Values shown are mean ± SEM (n = 2 and n = 4 mice for mock- and β-AS3-transduced samples, respectively). ns, not significant (unpaired t test). (D) Human hematopoietic progenitor content in BM MNC obtained from mice transplanted with mock- or β-AS3-transduced SCD Plerixafor-mobilized HSPCs. We plotted the percentage of BM MNC giving rise to BFU-E and CFU-GM. Values shown are mean ± SEM of 2 and 4 samples obtained from mice receiving mock- and β-AS3-transduced cells, respectively. (E) Average VCN/cell in pools of BFU-E and CFU-GM derived from the BM of mice injected with SCD Plerixafor-mobilized HSPCs transduced with β-AS3 (n = 4). Values shown are mean ± SEM. (F) Average VCN/cell in pools of BFU-E and CFU-GM obtained from mobilized HSPCs of a HD (mPB) and a SCD patient (SCD mPB) transduced with β-AS3 ± PGE2. Values shown are mean ± SEM of technical replicates.
We next evaluated β-AS3 gene transfer efficiency in long-term repopulating SCD mobilized HSCs. Mobilized HSPCs from an SCD patient were transduced at an MOI of 100, with a good manufacturing practices (GMPs)-engineering batch of β-AS3 LV, achieving an average VCN of 0.94 and 0.54 copies in in vitro-generated BFU-E and CFU-GM pools. Mock- and β-AS3-transduced cells were injected into 3 and 4 NSG mice, respectively. At 18 weeks after transplantation, we analyzed the engraftment of human hematopoietic cells and average VCN/cell. The engraftment of mock- and β-AS3-transduced cells was not statistically different in BM, spleen, and thymus (Figure 5C), with no skewing toward a particular lineage in any of the samples (Figure S5). Total human cells derived from the mouse BM were then cultured in a CFC assay. Mock- and β-AS3-transduced samples gave rise to a similar number of BFU-E and CFU-GM with an average VCN of 0.18 ± 0.05 and 0.16 ± 0.05, respectively (Figures 5D and 5E).
Finally, to increase gene transfer efficiency, we transduced HD and SCD mobilized HSPCs using a GMP-engineering batch of β-AS3 at an MOI of 100 in the presence of 16,16-dimethyl-prostaglandin E2 (PGE2).16 PGE2 treatment led to a 2- to 3.6-fold increase in VCN/cell, leading to 2.63 ± 1.07 and 1.52 ± 0.33 vector copies in BFU-E and CFU-GM derived from SCD Plerixafor-mobilized HSPCs (Figure 5F).
Discussion
In this study, we describe an advanced design of an anti-sickling HBB-expressing cassette in the context of an LV vector, to expedite high-titer vector production and maximize transduction efficiency of human HSPCs while guaranteeing high-level transgene expression for potential use in gene therapy for SCD. We compared two LVs containing an HS2 + HS3 or an HS2 + HS3 + HS4 mini-βLCR (β-AS3 and β-AS3 HS4), controlling the expression of a βAS3-globin transgene containing three anti-sickling amino acid substitutions.14
The β-AS3 LV showed a high transduction efficiency of HSPCs from SCD patients, leading to a potentially therapeutic production of βAS3-globin chains. Importantly, the high transduction efficiency of β-AS3 did not impair erythroid maturation, clonogenic potential, and in vivo engraftment of transduced HSPCs. A similar LV bearing only HS2 and HS3 sequences was shown to express therapeutic levels of a wild-type HBB transgene, thus correcting a murine model of β-thalassemia as well as human β-thalassemic HSPC-derived erythroblasts.17, 18 In line with these studies, preliminary results of a phase I/II gene therapy clinical trial treating β-thalassemia with this compact LV are very encouraging (G. Ferrari, 2016, Am. Soc. Hem., conference).12 However, the impact of the HS4 element in HBB-like-expressing LVs on transduction efficiency and transgene expression has not previously been reported, particularly in the context of SCD. Importantly, the correction of SCD requires high expression levels of the therapeutic HBB-like chain that needs to compete with the endogenous βS-globin for the incorporation into Hb tetramers, a situation that does not occur in β-thalassemia. Upon inclusion of a 1.1-kb HS4 element, LV transduction efficiency was severely affected, with a lower proportion of β-AS3 HS4 vector-positive cells and average transgene VCN per transduced cell compared to β-AS3 LV. In addition, in contrast to β-AS3, increasing doses of β-AS3 HS4 did not improve gene transfer efficiency, thereby indicating that the poor performance of the β-AS3-HS4 cannot be overcome by simply increasing MOI. The superior transduction potential of β-AS3 over β-AS3 HS4 was confirmed in vivo, where differences in gene marking were maintained in progenitors derived from long-term repopulating HSCs.
Premature truncation of the β-AS3 HS4 viral transcript occurred, due to a polyadenylation site present within the HS4 element. However, the production of full-length viral genomic RNAs was similar for β-AS3 HS4 and β-AS3 vectors, indicating that the generation of truncated transcripts is not the main determinant of the low β-AS3 HS4 LV titer. The sequence and/or the size of the 1.1-kb HS4 element may impair the production of a high-titer and infective LV by affecting the synthesis or the stability of ψ+ viral RNAs in the packaging cells. However, the use of a short 304-bp HS4 element restored the ψ+ viral RNA levels but did not significantly increase the viral titer and infectivity. Therefore, we hypothesize that the presence of HS4 sequences (e.g., the core HS4 element) might affect other steps of the viral life cycle, such as packaging of the viral RNAs, reverse transcription, or integration.
High levels of transgene expression are mandatory for a favorable outcome of gene therapy for SCD patients, where globin chain production from the transgene allows the displacement of the endogenous βS-globin chain from the hemoglobin tetramers, thus ameliorating the sickling RBC phenotype.13 Therefore, it was crucial to confirm that the removal of the HS4 element from the mini-LCR does not affect transgene expression, despite the beneficial enhancement in gene transfer efficiency. In transgenic mice harboring the human HBB locus, deletion of the 280-bp core element of HS4 leads to a severe HBB downregulation,19 but deletion of the entire HS4 has a mild effect on HBB expression.20 Interestingly, we show that the HS4 element is dispensable for boosting HBB transgene expression in the context of the β-AS3 LV in primary human erythroblasts. Indeed, we detected comparable levels of βAS3-globin mRNA and HbAS3 tetramers in erythroid cells derived from samples harboring the same β-AS3 or β-AS3 HS4 average VCN per genome. In another HBB-expressing LV, the truncation of the 5′ region of the βLCR HS4 element was associated with a higher gene marking in a murine model of β-thalassemia, but lower transgene expression levels, as compared to the LV containing the full HS4.21 This discrepancy with our study may be ascribed to the potential different activity of HS4 in murine and human erythroid cells. Of note, our vector contains a larger HS2 element (∼1.5 kb versus 0.8 kb21) that in combination with only HS3 may be sufficient per se to produce high HBB transgene expression levels. Importantly, we observed a linear correlation between the vector copies and HBB transgene expression for both β-AS3 HS4 and β-AS3 LVs, suggesting that HS4 element is dispensable for integration site position-independent transgene expression.
The remarkable transduction efficiency of SCD HSPCs achieved with β-AS3 combined with the high βAS3-globin expression levels allowed accumulation of up to 60% therapeutic hemoglobin in terminally differentiated RBCs, levels that were never achieved with the β-AS3 HS4 LV even at high MOI. A concomitant decrease of HbS tetramers was observed, due to the competition of the βAS3-globin chain for incorporation into Hb tetramers, likely favored by the presence of the A22 amino acidic residue that increases the affinity of the βAS3-globin for HBA.14 The sustained anti-sickling βAS3-globin production (27%–47% of HBAS3) together with the beneficial reduction of HbS levels were able to inhibit the formation of hemoglobin polymers and reduce RBC sickling (34%–47% of corrected RBCs), proving the efficacy of the β-AS3 LV to reverse the major cellular manifestation of SCD. Of note, in compound heterozygotes of SCD and HPFH, a condition characterized by maintenance of elevated amounts of anti-sickling fetal HBG polypeptides in adult life, HbF levels accounting for as little as 20%–30% of the total hemoglobin are associated with the absence of SCD symptoms.22
Compared to the BB305 vector, which has given encouraging results in β-thalassaemic patients23 but not consistently in those with SCD (J. Kanter et al., 2011, Am. Soc. Hem., conference),13 β-AS3 differs in two key components. First, the mini-HBB in β-AS3 contains three anti-sickling amino acid substitutions (G16D, E22A, T87Q) compared to one (T87Q) in BB305,13 giving β-AS3 a greater anti-sickling activity. Second, the mini-βLCR in BB305 consists of the HS2, HS3, and HS4 elements but is only 2.6 kb in length, whereas the mini-βLCR in β-AS3, which, although it consists of just the HS2 and HS3 sites, spans 2.7 kb. Thus, the HS core regions in β-AS3 possess considerably larger flanking sequences compared to those in BB305. Given that distance between the core regions of the βLCR HS sites is crucial for effective mutual interaction and chromatin looping leading to βLCR engagement on a downstream promoter for efficient transcriptional activation,7 the greater compactness of the βLCR HS sites in BB305 could be a disadvantage.
Studies in SCD patients with mixed-donor chimerism following allogeneic transplantation indicate that 20% of corrected HSCs are sufficient to reverse the sickle phenotype,24, 25, 26 in all likelihood due to the increased half-life of corrected over unmodified RBCs in vivo. In this regard, it is noteworthy that we achieved ∼20% of transduced long-term repopulating HSCs after transplantation of HSPCs derived from SCD patients either harvested from BM or mobilized using Plerixafor.15 Plerixafor-mobilized SCD HSPCs represent the only available source of HSCs for a gene therapy approach to SCD, since the administration of the conventional mobilizing agent G-CSF in individuals with SCD leads to severe adverse events,27, 28, 29 and the recovery of HSPCs from SCD patients’ BM is peculiarly low (M.C., unpublished data). Importantly, exposure to PGE2, a known molecule that increases gene transfer in long-term repopulating stem cells,16 enhanced transduction efficiency in SCD Plerixafor-mobilized HSPCs.
In conclusion, in this study we report the functional characterization of a novel and efficient LV expressing an anti-sickling HBB transgene in clinically relevant cells from SCD patients as part of a program of work aimed at clinical translation of an effective LV-based gene therapy approach for SCD. Thus, findings of good transduction and expression efficiencies of mobilized HSPCs from SCD patients with the β-AS3 LV bodes well for consistent efficacious future clinical application.
Materials and Methods
Vector Construction
The expression cassette consisting of DNase I HSs HS2 (genomic coordinates[hg38], chr11:5280255-5281665) and HS3 (genomic coordinates[hg38], chr11:5284251-5285452) of the βLCR30 and HBB mini-gene extending from −265 bp upstream of the transcriptional start site to +300 bp downstream of the poly(A)-addition site (genomic coordinates[hg38], chr11:5225174-5227336) with a short version of intron 2 (genomic coordinates[hg38], chr11:5203703-5204295)17 was cloned into a pCCL LV backbone31 to generate the pCCL.β-globin plasmid. The mutations determining three anti-sickling amino acid substitutions (G16D, E22A, and T87Q)14 were introduced in the pCCL.β-globin plasmid by in vitro site-directed mutagenesis to obtain the pCCL.β-AS3 plasmid. The addition of the βLCR HS4 element (genomic coordinates[hg38], chr11:5287846-5288936)30 or of the short HS4 fragment (genomic coordinates[hg38], chr11:5288094-5288397) containing the core HS4 element to generate the pCCL.β-AS3 HS4 and the pCCL.β-AS3 HS4 core plasmids was achieved by replacing the HS2 and HS3 sequence of pCCL.β-AS3 with a commercially synthesized (GeneWiz, South Plainfield, NJ, USA) fragment containing a combination of βLCR HS2, HS3, and HS4 sites as previously described.32
LV Production and Titration
Third-generation LVs were produced by calcium phosphate transient transfection of HEK293T cells of the transfer vector (pCCL.β-AS3 or pCCL.β-AS3 HS4), the packaging plasmid pKLg/p.RRE, the Rev-encoding plasmid pK.REV, and the vesicular stomatitis VSV-G envelope-encoding plasmid pK.G, as previously described.33 The physical titer of vector preparations was measured using the HIV-1 Gag p24 antigen immunocapture assay kit (PerkinElmer, Waltham, MA, USA) and expressed as p24 ng/mL. The viral infectious titer was calculated by transducing HCT116, K562, and HEL cells. HCT116 cells were transduced with serial vector dilutions, as previously described.34 K562 and HEL cells were transduced overnight with serial dilutions of the virus in DMEM supplemented of 10% fetal bovine serum (Gibco, Carlsbad, CA, USA), penicillin and streptomycin (Gibco, Carlsbad, CA, USA), glutamine (Gibco, Carlsbad, CA, USA), and polybrene (8 μg/mL, Sigma-Aldrich, St. Louis, MI, USA). After transduction, cells were washed and maintained in complete DMEM for 1 week before determining VCN per genome. VCN was used to calculate the viral infectious titer, expressed as transducing units per mL (TU/mL). Viral infectivity was calculated as ratio between infectious and physical titers (TU/ng p24).
Viral Transcript Quantification
Total RNA was extracted from HEK293T cells transfected as described above (“Viral vector preparation”), using the RNeasy kit (QIAGEN, Venlo, Netherlands), which included an in-column DNase I treatment step. An additional DNase treatment was performed after RNA purification using the TURBO DNA-free kit (Ambion, Waltham, MA, USA). Then, RNA was reverse-transcribed by extension of random hexamers using the SuperScript III First-Strand Synthesis kit (Invitrogen, Carlsbad, CA, USA). As negative controls, RNA samples were treated without reverse transcriptase (RT). We performed duplex TaqMan qRT-PCR to quantify total ψ+ and full-length viral transcripts. GAPDH was used as an internal reference standard for RNA normalization.
Primers and probes used for TaqMan qRT-PCR are as follows:
HIV-1-RNAfull-length 3 (ΔU3) FOR 5′-CAGCTGTAGATCTTAGCCACTTT-3′
HIV-1-RNAfull-length (ΔU3) REV 5′-AGCAGATCTTGTCTTCGTTGG-3′
HIV-1-RNAfull-length (ΔU3) PROBE-fam AS 5′-AGTGAATTAGCCCTTCCAGTCCC-3′
HIV-1-PSI FOR 5′-CAGGACTCGGCTTGCTGAAG-3′
HIV-1-PSI REV 5′-TCCCCCGCTTAATACTGACG-3′
HIV-1-PSI PROBE-fam 5′-CGCACGGCAAGAGGCGAGG-3′
GAPDH FOR 5′-CTTCATTGACCTCAACTACATGGTTT-3′
GAPDH REV 5′-TGGGATTTCCATTGATGACAAG-3′
GAPDH PROBE-vic 5′-CAAATTCCATGGCACCGTCAAGGC-3′.
Identification of Truncated Viral Transcripts
Total RNA was extracted from packaging HEK293T cells following transfection for vector packaging and reverse-transcribed by using the 3′ RACE Kit (Invitrogen, Carlsbad, CA, USA), in accordance with the manufacturer’s instructions. To identify any transcripts with premature polyadenylation within the HS4 region, we used a forward primer annealing to the HS3 region (HS3 primer 5′-CTCTTACTCATCCCATCACGTATGC-3′) and the UAP primer provided in the kit as reverse primer (5′-CUACUACUACUAGGCCACGCGTCGACTAGTAC-3′). The 3′ RACE PCR product was loaded on a 1% agarose gel, and the amplicons were visualized by GelRed (Biotium, Fremont, CA, USA) staining. After electrophoresis, the amplicons were extracted from the gel, cloned using the StrataClone PCR Cloning Kit (Agilent, Santa Clara, CA, USA), and analyzed by Sanger sequencing.
HSPC Purification and Transduction
We obtained human adult CD34+ HSPCs mobilized with G-CSF from HDs. Human adult SCD HSPCs were either harvested from BM or mobilized using Plerixaflor (NCT 02212535 clinical trial, Necker Hospital, Paris, France). Written informed consent was obtained from all subjects. All experiments were performed in accordance with the Declaration of Helsinki. The study was approved by the regional investigational review board (reference, DC 2014-2272, CPP Ile-de-France II “Hôpital Necker-Enfants malades”). HSPCs were purified by immunomagnetic selection with AutoMACS (Miltenyi Biotec, Bergisch Gladbach, Germany) after immunostaining with CD34 MicroBead Kit (Miltenyi Biotec, Bergisch Gladbach, Germany).
CD34+ cells (106 cells/mL) were pre-activated for 24 hr in X-VIVO 20 supplemented with penicillin and streptomycin (Gibco, Carlsbad, CA, USA) and the following recombinant human cytokines (obtained from either CellGenix, Freiburg, Germany or Peprotech, London, UK): 300 ng/mL SCF, 300 ng/mL Flt-3L, 100 ng/mL TPO, and 20 ng/mL interleukin-3 (IL-3). After pre-activation, cells (106 cells/mL) were transduced for 24 hr in plates coated with RetroNectin (10 μg/cm2, Takara Bio, Kusatsu, Japan), in the pre-activation medium supplemented with protamine sulfate (4 μg/mL, Sigma-Aldrich, St. Louis, MI, USA or APP Pharmaceuticals, Schaumburg, IL, USA). Transductions using PGE2 (Cayman Chemical) efficacy were performed using conditions previously described.13, 16 MOI was calculated using the viral infectious titer determined in HCT116 cells.
In Vitro Erythroid Differentiation
A three-step protocol35 was adapted to generate mature RBCs from mock- and LV-transduced CD34+ cells. From days 0 to 6, cells were grown in a basal erythroid medium supplemented with the following recombinant human cytokines: 100 ng/mL SCF (Peprotech, London, UK), 5 ng/mL IL3 (Peprotech, London, UK), and 3 IU/mL of erythropoietin (EPO) Eprex (Janssen-Cilag, Issy-Les-Moulineaux, France) and hydrocortisone (Sigma, St. Louis, MI, USA) at 10−6 M. From days 6 to 9, cells were cultured onto a layer of murine stromal MS-5 cells in basal erythroid medium supplemented only with 3 IU/mL EPO Eprex. Finally, from days 9 to 20, cells were continued to be cultured on a layer of MS-5 cells in basal erythroid medium but without cytokines. Erythroid differentiation was monitored by May Grunwald-Giemsa staining, flow cytometry analysis of the erythroid surface markers CD36 (CD36-V450, BD Horizon, Franklin Lakes, NJ, USA), CD71 (CD71-FITC, BD PharMingen, Franklin Lakes, NJ, USA) and glyophorin A (GYPA) (CD235a-PECY7, BD PharMingen, Franklin Lakes, NJ, USA). We used the nuclear dye DRAQ5 (eBioscience, San Diego, CA, USA) to evaluate the proportion of enucleated RBCs. Flow cytometry analyses were performed using the Gallios analyzer and Kaluza software (Beckman-Coulter, Brea, CA, USA).
CFC Assay
The number of hematopoietic progenitors was evaluated by a clonal colony-forming cell (CFC) assay, where HSPCs were plated at 1 × 103 cells/mL in methylcellulose-containing medium (GFH4435, Stem Cell Technologies, Vancouver, BC, USA) under conditions supporting both erythroid and granulo-monocytic differentiation. Numbers of BFU-E and CFU-GM colonies were scored after 14 days. BFU-E were randomly picked and collected as bulk populations (containing at least 25 colonies) or individual BFU-E to evaluate the efficiency of transduction and hemoglobin expression.
VCN Analysis
At day 13 of erythroid differentiation, or 14 days after plating HSPCs in the methylcellulose medium, DNA was extracted from transduced cells using the ReliaPrep Blood gDNA Miniprep System (Promega, Madison, WI, USA) or the DNA Extract All Reagents Kit (Thermo Fisher Scientific, Waltham, MA, USA). VCN per diploid genome was determined by duplex TaqMan qPCR (Applied Biosystems, Foster City, CA, USA), as previously described.34 We used primers and probes specific for (1) the viral ψ (PSI) packaging signal (HIV-1-PSI FOR 5′-CAGGACTCGGCTTGCTGAAG-3′, HIV-1-PSI REV 5′-TCCCCCGCTTAATACTGACG-3′, HIV-1-PSI PROBE 5′-CGCACGGCAAGAGGCGAGG-3′) and (2) the human albumin gene (ALB) as an internal reference standard (ALB FOR 5′-GCTGTCATCTCTTGTGGGCTGT-3′, ALB REV 5′-ACTCATGGGAGCTGCTGGTTC-3′, ALB PROBE 5′-CCTGTCATGCCCACACAAATCTCTCC-3′). Standard curves were obtained by serial dilutions of a plasmid containing one copy of ψ and ALB sequences. The number of PSI and ALB copies in test samples was extrapolated from the standard curves.
qRT-PCR Analysis of Globin Transcripts
At day 13 of erythroid differentiation, or 14 days after plating HSPCs in the methylcellulose medium, RNA was extracted using RNeasy micro kit (QIAGEN, Venlo, Netherlands). Reverse transcription of mRNA employed the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA, USA) with oligo(dT) primers. qRT-PCR was performed using SYBR green (Applied Biosystems, Foster City, CA, USA) as a detection system with the following primers:
HBBAS3 F: 5′-AAGGGCACCTTTGCCCAG-3′
HBBAS3 R: 5′-GCCACCACTTTCTGATAGGCAG-3′
HBB F: 5′-AAGGGCACCTTTGCCACA-3′
HBB R: 5′-GCCACCACTTTCTGATAGGCAG-3′.
Cation-Exchange HPLC Analysis of Hemoglobin Tetramers
Cation-exchange HPLC analysis was performed using a NexeraX2 SIL-30AC chromatograph (Shimadzu, Kyoto, Japan) and the LC Solution software. Hemoglobin tetramers from mature RBCs or BFU-E were separated by HPLC using a 2 cation-exchange column (PolyCAT A, PolyLC, Columbia, MD, USA). Samples were eluted with a gradient mixture of solution A (20 mM bis Tris, 2 mM KCN, pH, 6.5) and solution B (20 mM bis Tris, 2 mM KCN, 250 mM NaCl, pH, 6.8). The absorbance was measured at 415 nm.
Sickling Assay
RBCs at day 20 of erythroid differentiation, derived from transduced and mock-transduced SCD HSPCs, were exposed to an oxygen-deprived atmosphere (0% O2) for at least 35 min before image capture, using the AxioObserver Z1 microscope (Zeiss, Oberkochen, Germany) and a 20× objective. Images were processed with ImageJ to determine the percentage of sickled RBCs per field of acquisition in the total RBC population.
NSG Mouse Transplantation
Non-obese diabetic severe combined immunodeficiency gamma (NSG) mice (NOD.CgPrkdcscid Il2rgtm1Wj/SzJ, Charles River Laboratories, St Germain sur l’Arbresle, France) were housed in a specific pathogen-free facility. Mice at 6 to 8 weeks of age were conditioned with busulfan (Sigma, St Louis, MO, USA) injected intraperitoneally (25 mg/kg body weight/day) 24 hr, 48 hr, and 72 hr before transplantation. Mock- or LV-transduced CD34+ cells (500,000 cells/mouse for SCD BM, HD mPB, and HD CB HSPCs and 106 cells/mouse for SCD-mobilized HSPCs) were transplanted into NSG mice via retro-orbital sinus injection. Neomycin and acid water were added in the water bottle. At 10 to 19 weeks post-transplantation, NSG recipients were sacrificed. Cells were harvested from femur BM, thymus, and spleen, stained with antibodies against murine or human surface markers (murine CD45, BD Biosciences, Franklin Lakes, NJ, USA; human CD45, Miltenyi Biotec, Bergisch Gladbach, Germany; human CD3, Miltenyi Biotec, Bergisch Gladbach, Germany; human CD14, BD Biosciences, Franklin Lakes, NJ, USA; human CD15, Beckman Coulter, Brea, CA, USA; human CD19, Sony Biotechnologies, San Jose, CA, USA; human CD36, BD Biosciences, Franklin Lakes, NJ, USA), and analyzed by flow cytometry using a Gallios analyzer and the Kaluza software (Beckman-Coulter, Brea, CA, USA). 100,000 BM cells were subjected to the CFC assay, and BFU-E and CFU-GM pools were analyzed for VCN. All experiments and procedures were performed in compliance with the French Ministry of Agriculture’s regulations on animal experiments and were approved by the regional Animal Care and Use Committee (APAFIS#2101-2015090411495178 v4).
Statistical Analyses
Unpaired t tests were performed to compare vector titer and infectivity, viral genomic RNA levels, VCN/cell and the engraftment rate between 2 groups. We used one-way ANOVA test when comparing the engraftment rate between >2 groups. We performed chi-square test (two-sample test for equality of proportions with continuity correction) to compare the frequency of transduced CFCs. Linear regression analyses were performed to assess the correlation between VCN/cell and transgene expression. All statistical analyses were performed using Prism4 software (GraphPad, La Jolla, CA, USA). The threshold for statistical significance was set to p < 0.05.
Author Contributions
L.W. and V.P. designed and conducted experiments and wrote the paper; E.M., C.A., S.M., and V.M. designed and conducted experiments; C.B., H.S., and T.F. conducted experiments; W.E.-N. contributed to the design of the experimental strategy; F.M., M.C., and I.A.-S. conceived the study; M.N.A. assisted with the drafting of the manuscript; A.M. conceived the study, designed experiments, and wrote the paper.
Conflicts of Interest
The authors have no conflict of interest.
Acknowledgments
This work was supported by grants from AFM-Telethon (17224), the European Research Council (ERC-2010-AdG, GT-SKIN; ERC-2015-AdG, GENEFORCURE), the Agence Nationale de la Recherche (ANR-16-CE18-0004 and ANR-10-IAHU-01 “Investissements d’avenir” program), the EU Marie Curie-COFUND(PRESTIGE_2015_2_0015) program, and a collaboration with BioMarin Pharmaceutical Inc.
Footnotes
Supplemental Information includes five figures and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.07.012.
Supplemental Information
References
- 1.Piel F.B., Patil A.P., Howes R.E., Nyangiri O.A., Gething P.W., Dewi M., Temperley W.H., Williams T.N., Weatherall D.J., Hay S.I. Global epidemiology of sickle haemoglobin in neonates: a contemporary geostatistical model-based map and population estimates. Lancet. 2013;381:142–151. doi: 10.1016/S0140-6736(12)61229-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinberg M.H., Barton F., Castro O., Pegelow C.H., Ballas S.K., Kutlar A., Orringer E., Bellevue R., Olivieri N., Eckman J. Effect of hydroxyurea on mortality and morbidity in adult sickle cell anemia: risks and benefits up to 9 years of treatment. JAMA. 2003;289:1645–1651. doi: 10.1001/jama.289.13.1645. [DOI] [PubMed] [Google Scholar]
- 3.Chandrakasan S., Malik P. Gene therapy for hemoglobinopathies: the state of the field and the future. Hematol. Oncol. Clin. North Am. 2014;28:199–216. doi: 10.1016/j.hoc.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pawliuk R., Westerman K.A., Fabry M.E., Payen E., Tighe R., Bouhassira E.E., Acharya S.A., Ellis J., London I.M., Eaves C.J. Correction of sickle cell disease in transgenic mouse models by gene therapy. Science. 2001;294:2368–2371. doi: 10.1126/science.1065806. [DOI] [PubMed] [Google Scholar]
- 5.Perumbeti A., Higashimoto T., Urbinati F., Franco R., Meiselman H.J., Witte D., Malik P. A novel human gamma-globin gene vector for genetic correction of sickle cell anemia in a humanized sickle mouse model: critical determinants for successful correction. Blood. 2009;114:1174–1185. doi: 10.1182/blood-2009-01-201863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero Z., Urbinati F., Geiger S., Cooper A.R., Wherley J., Kaufman M.L., Hollis R.P., de Assin R.R., Senadheera S., Sahagian A. β-globin gene transfer to human bone marrow for sickle cell disease. J. Clin. Invest. 2013:67930. doi: 10.1172/JCI67930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim A., Dean A. Chromatin loop formation in the β-globin locus and its role in globin gene transcription. Mol. Cells. 2012;34:1–5. doi: 10.1007/s10059-012-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.May C., Rivella S., Callegari J., Heller G., Gaensler K.M., Luzzatto L., Sadelain M. Therapeutic haemoglobin synthesis in beta-thalassaemic mice expressing lentivirus-encoded human beta-globin. Nature. 2000;406:82–86. doi: 10.1038/35017565. [DOI] [PubMed] [Google Scholar]
- 9.Persons D.A., Hargrove P.W., Allay E.R., Hanawa H., Nienhuis A.W. The degree of phenotypic correction of murine beta -thalassemia intermedia following lentiviral-mediated transfer of a human gamma-globin gene is influenced by chromosomal position effects and vector copy number. Blood. 2003;101:2175–2183. doi: 10.1182/blood-2002-07-2211. [DOI] [PubMed] [Google Scholar]
- 10.Molete J.M., Petrykowska H., Bouhassira E.E., Feng Y.Q., Miller W., Hardison R.C. Sequences flanking hypersensitive sites of the beta-globin locus control region are required for synergistic enhancement. Mol. Cell. Biol. 2001;21:2969–2980. doi: 10.1128/MCB.21.9.2969-2980.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavazzana M., Antoniani C., Miccio A. Gene Therapy for β-Hemoglobinopathies. Mol. Ther. 2017;25:1142–1154. doi: 10.1016/j.ymthe.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrari G., Cavazzana M., Mavilio F. Gene Therapy Approaches to Hemoglobinopathies. Hematol. Oncol. Clin. North Am. 2017;31:835–852. doi: 10.1016/j.hoc.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Ribeil J.A., Hacein-Bey-Abina S., Payen E., Magnani A., Semeraro M., Magrin E., Caccavelli L., Neven B., Bourget P., El Nemer W. Gene Therapy in a Patient with Sickle Cell Disease. N. Engl. J. Med. 2017;376:848–855. doi: 10.1056/NEJMoa1609677. [DOI] [PubMed] [Google Scholar]
- 14.Levasseur D.N., Ryan T.M., Reilly M.P., McCune S.L., Asakura T., Townes T.M. A recombinant human hemoglobin with anti-sickling properties greater than fetal hemoglobin. J. Biol. Chem. 2004;279:27518–27524. doi: 10.1074/jbc.M402578200. [DOI] [PubMed] [Google Scholar]
- 15.Lagresle-Peyrou C., Lefrère F., Magrin E., Ribeil J.A., Romano O., Weber L., Magnani A., Sadek H., Plantier C., Gabrion A. Plerixafor enables safe, rapid, efficient mobilization of hematopoietic stem cells in sickle cell disease patients after exchange transfusion. Haematologica. 2018;103:778–786. doi: 10.3324/haematol.2017.184788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zonari E., Desantis G., Petrillo C., Boccalatte F.E., Lidonnici M.R., Kajaste-Rudnitski A., Aiuti A., Ferrari G., Naldini L., Gentner B. Efficient Ex Vivo Engineering and Expansion of Highly Purified Human Hematopoietic Stem and Progenitor Cell Populations for Gene Therapy. Stem Cell Reports. 2017;8:977–990. doi: 10.1016/j.stemcr.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miccio A., Cesari R., Lotti F., Rossi C., Sanvito F., Ponzoni M., Routledge S.J., Chow C.M., Antoniou M.N., Ferrari G. In vivo selection of genetically modified erythroblastic progenitors leads to long-term correction of beta-thalassemia. Proc. Natl. Acad. Sci. USA. 2008;105:10547–10552. doi: 10.1073/pnas.0711666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roselli E.A., Mezzadra R., Frittoli M.C., Maruggi G., Biral E., Mavilio F., Mastropietro F., Amato A., Tonon G., Refaldi C. Correction of beta-thalassemia major by gene transfer in haematopoietic progenitors of pediatric patients. EMBO Mol. Med. 2010;2:315–328. doi: 10.1002/emmm.201000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navas P.A., Peterson K.R., Li Q., McArthur M., Stamatoyannopoulos G. The 5'HS4 core element of the human beta-globin locus control region is required for high-level globin gene expression in definitive but not in primitive erythropoiesis. J. Mol. Biol. 2001;312:17–26. doi: 10.1006/jmbi.2001.4939. [DOI] [PubMed] [Google Scholar]
- 20.Fedosyuk H., Peterson K.R. Deletion of the human beta-globin LCR 5‘HS4 or 5’HS1 differentially affects beta-like globin gene expression in beta-YAC transgenic mice. Blood Cells Mol. Dis. 2007;39:44–55. doi: 10.1016/j.bcmd.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lisowski L., Sadelain M. Locus control region elements HS1 and HS4 enhance the therapeutic efficacy of globin gene transfer in beta-thalassemic mice. Blood. 2007;110:4175–4178. doi: 10.1182/blood-2007-08-108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ngo D.A., Aygun B., Akinsheye I., Hankins J.S., Bhan I., Luo H.Y., Steinberg M.H., Chui D.H. Fetal haemoglobin levels and haematological characteristics of compound heterozygotes for haemoglobin S and deletional hereditary persistence of fetal haemoglobin. Br. J. Haematol. 2012;156:259–264. doi: 10.1111/j.1365-2141.2011.08916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson A.A., Walters M.C., Kwiatkowski J., Rasko J.E.J., Ribeil J.A., Hongeng S., Magrin E., Schiller G.J., Payen E., Semeraro M. Gene Therapy in Patients with Transfusion-Dependent β-Thalassemia. N. Engl. J. Med. 2018;378:1479–1493. doi: 10.1056/NEJMoa1705342. [DOI] [PubMed] [Google Scholar]
- 24.Altrock P.M., Brendel C., Renella R., Orkin S.H., Williams D.A., Michor F. Mathematical modeling of erythrocyte chimerism informs genetic intervention strategies for sickle cell disease. Am. J. Hematol. 2016;91:931–937. doi: 10.1002/ajh.24449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walters M.C., Patience M., Leisenring W., Rogers Z.R., Aquino V.M., Buchanan G.R., Roberts I.A., Yeager A.M., Hsu L., Adamkiewicz T., Multicenter Investigation of Bone Marrow Transplantation for Sickle Cell Disease Stable mixed hematopoietic chimerism after bone marrow transplantation for sickle cell anemia. Biol. Blood Marrow Transplant. 2001;7:665–673. doi: 10.1053/bbmt.2001.v7.pm11787529. [DOI] [PubMed] [Google Scholar]
- 26.Abraham A., Hsieh M., Eapen M., Fitzhugh C., Carreras J., Keesler D., Guilcher G., Kamani N., Walters M.C., Boelens J.J., National Institutes of Health. Center for International Blood and Marrow Transplant Research Relationship between Mixed Donor-Recipient Chimerism and Disease Recurrence after Hematopoietic Cell Transplantation for Sickle Cell Disease. Biol. Blood Marrow Transplant. 2017;23:2178–2183. doi: 10.1016/j.bbmt.2017.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abboud M., Laver J., Blau C.A. Granulocytosis causing sickle-cell crisis. Lancet. 1998;351:959. doi: 10.1016/S0140-6736(05)60614-9. [DOI] [PubMed] [Google Scholar]
- 28.Adler B.K., Salzman D.E., Carabasi M.H., Vaughan W.P., Reddy V.V., Prchal J.T. Fatal sickle cell crisis after granulocyte colony-stimulating factor administration. Blood. 2001;97:3313–3314. doi: 10.1182/blood.v97.10.3313. [DOI] [PubMed] [Google Scholar]
- 29.Grigg A.P. Granulocyte colony-stimulating factor-induced sickle cell crisis and multiorgan dysfunction in a patient with compound heterozygous sickle cell/beta+ thalassemia. Blood. 2001;97:3998–3999. doi: 10.1182/blood.v97.12.3998. [DOI] [PubMed] [Google Scholar]
- 30.Hardison R., Slightom J.L., Gumucio D.L., Goodman M., Stojanovic N., Miller W. Locus control regions of mammalian beta-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- 31.Follenzi A., Sabatino G., Lombardo A., Boccaccio C., Naldini L. Efficient gene delivery and targeted expression to hepatocytes in vivo by improved lentiviral vectors. Hum. Gene Ther. 2002;13:243–260. doi: 10.1089/10430340252769770. [DOI] [PubMed] [Google Scholar]
- 32.Montiel-Equihua C.A., Zhang L., Knight S., Saadeh H., Scholz S., Carmo M., Alonso-Ferrero M.E., Blundell M.P., Monkeviciute A., Schulz R. The β-globin locus control region in combination with the EF1α short promoter allows enhanced lentiviral vector-mediated erythroid gene expression with conserved multilineage activity. Mol. Ther. 2012;20:1400–1409. doi: 10.1038/mt.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cantore A., Ranzani M., Bartholomae C.C., Volpin M., Valle P.D., Sanvito F., Sergi L.S., Gallina P., Benedicenti F., Bellinger D. Liver-directed lentiviral gene therapy in a dog model of hemophilia B. Sci. Transl. Med. 2015;7:277ra28. doi: 10.1126/scitranslmed.aaa1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lattanzi A., Duguez S., Moiani A., Izmiryan A., Barbon E., Martin S., Mamchaoui K., Mouly V., Bernardi F., Mavilio F., Bovolenta M. Correction of the Exon 2 Duplication in DMD Myoblasts by a Single CRISPR/Cas9 System. Mol. Ther. Nucleic Acids. 2017;7:11–19. doi: 10.1016/j.omtn.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giarratana M.C., Kobari L., Lapillonne H., Chalmers D., Kiger L., Cynober T., Marden M.C., Wajcman H., Douay L. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat. Biotechnol. 2005;23:69–74. doi: 10.1038/nbt1047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.