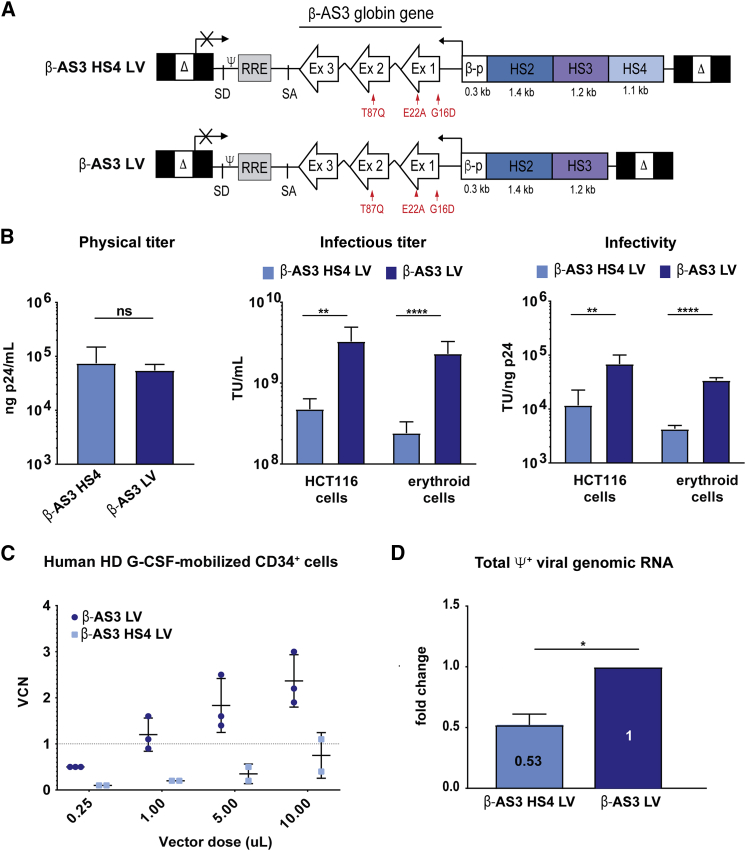
Figure 1.
Characterization of β-AS3 HBB-Expressing LVs
(A) Schematic representation of β-AS3 HS4 and β-AS3 lentiviral vectors. Δ, deleted HIV-1 U3 region; SD and SA, HIV splicing donor and acceptor sites; ψ, HIV-1 packaging signal; RRE, HIV-1 Rev responsive element; Ex, exons of the human HBB; βp, promoter of HBB; HS2, 3, and 4, DNase I hypersensitive site 2, 3, and 4 of human HBB LCR; red arrows indicate the mutations introduced in exon 1 (generating amino acid substitutions G16D and E22A) and exon 2 (generating amino acid substitution T87Q). (B) The histograms show the physical and infectious titers and infectivity of β-AS3 HS4 and β-AS3 LVs. Infectious titer and infectivity were measured in HTC116 (five different preparations for each vector) and K562 and HEL erythroid cell lines (two viral preparations per vector). (C) Vector copy number (VCN) in G-CSF-mobilized CD34+ cells from healthy donors (HDs). HSPCs were transduced with increasing amounts of three and two preparations of β-AS3 and β-AS3 HS4 LVs, respectively. Cells were grown in liquid culture, and after 1 week, VCN was determined. A linear correlation between β-AS3 vector dose and VCN is obtained, whereas a modest increase in VCN was achieved by transducing HSPCs with higher amounts of β-AS3 HS4 vector preparations. (D) Relative quantification of the ψ+ viral genomic RNA produced by HEK293T packaging cells (n = 3) by qRT-PCR. The β-AS3 sample was used as a calibrator. Data were plotted as mean ± SD (unpaired t test). *p ≤ 0.05, **p ≤ 0.01, ****p ≤ 0.0001, ns, not significant.