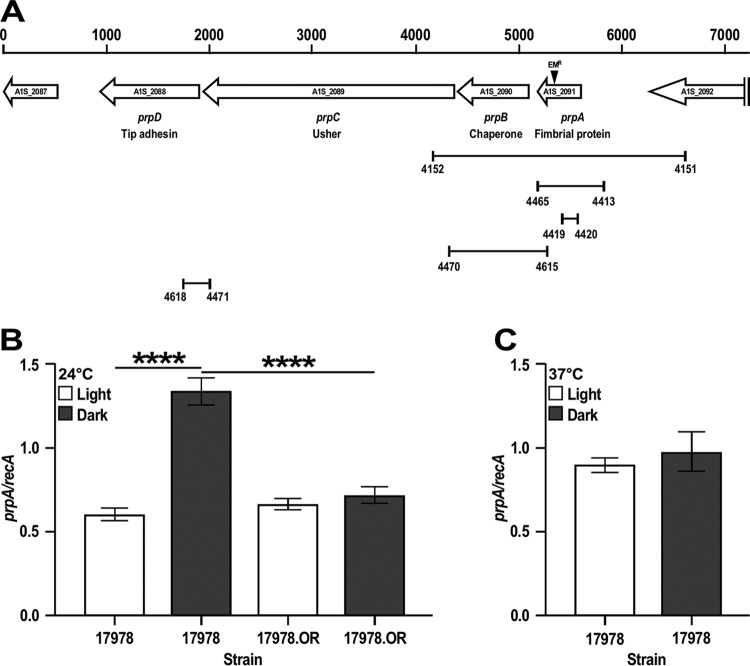
FIG 1.
Genetic organization and expression analysis of the prpABCD operon. (A) In silico analysis of the A1S_2089-A1S_2091 coding region. The horizontal arrows represent predicted coding regions and their direction of transcription. Coding regions are identified by their original genomic annotation, the names assigned in this work, and their predicted protein products. Numbers on the top of the vertical bars represent size in base pairs. The location of the insertion of the Emr DNA cassette within prpA is indicated by the black triangle. The connected vertical bars indicate primer locations and the lengths of cognate amplicons used to clone and mutagenize prpA, construct the complementing vector pMU1269 (Table 1), test the expression of prpA by qRT-PCR, and determine the polycistronic nature of the prpABCD operon. Numbers underneath the vertical bars identify primers listed in Table S2 in the supplemental material. (B) qRT-PCR of prpA using RNA isolated from 17978 and 17978.OR cells cultured in SB at 24°C under darkness or illumination. (C) qRT-PCR of prpA using RNA isolated from 17978 cells cultured in SB at 37°C under darkness or illumination. recA was used as a constitutively expressed control gene. Horizontal bars identify statistically different values (****, P ≤ 0.0001), and error bars represent the standard errors of each data set.