Fig. 2.
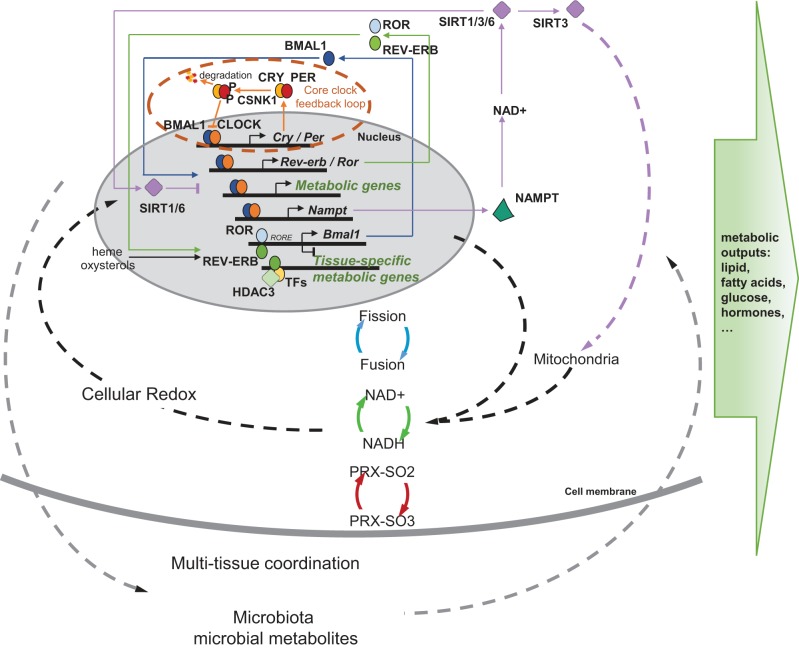
A multilayer molecular network couples the circadian clock to metabolic functions. The core circadian clock machinery consists of a transcriptional and translational feedback loop, in which transcription of the repressors (PER1/2/3 and CRY1/2) is activated by the transcriptional activators (CLOCK and BMAL1) and is turned off as the repressor proteins accumulate and suppress the activity of the CLOCK-BMAL1 heterodimers. CSNK1 phosphorylates the PER proteins to reregulate PER nuclear entry and turnover, controlling the velocity of the clock. This transcriptional/translational clock machinery produces a cyclic transcriptional profile of metabolic genes and is coupled with metabolic functions via multiple mechanisms. The clock is stabilized by a feedback loop involving nuclear receptors RORs and REV-ERBs, which is sensitive to cellular metabolic state and is involved in the transcriptional programming of tissue-specific metabolic output. The clock also directs NAD+ biosynthesis, which, in turn, negatively feeds back on CLOCK-BMAL1 transcriptional activity through the NAD+-dependent deacetylases (SIRTs). SIRT1 and SIRT6 are involved in the epigenetic regulation of metabolic genes, and SIRT3 regulates mitochondrial activity. Through this link and many others, the clock is coupled with daily fluctuations of the cellular redox state, which involves mitochondrial fission and fusion, NAD+/NADH flux, and peroxiredoxin hyperoxidation (PRX-SO2/3). These cellular circadian-metabolic mechanisms are also coordinated at the whole organism level among multiple tissues and interact with the microbiota to shape the overall profile of metabolic health.