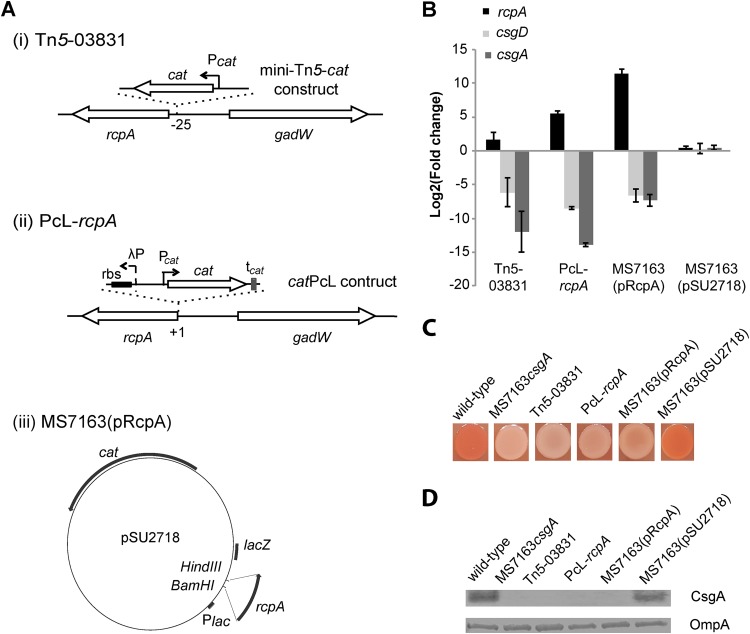
FIG 5 .
Overexpression of RcpA abolishes curli production. (A) Schematic overview of genetic constructions in rcpA-overexpressing mutants. (i) Tn5-03831 mutant, containing the mini-Tn5 at nucleotide −25 relative to the rcpA predicted ATG start codon (Pcat; cat gene promoter); (ii) PcL-rcpA mutant, PcL promoter and optimized ribosome binding site (rbs) inserted upstream of the rcpA CDS; and (iii) plasmid pRcpA, which contains the rcpA CDS cloned behind the lac promoter in the expression vector pSU2718. (B) Relative fold change in transcript level of rcpA, csgD, and csgA in rcpA-overexpressing mutants compared to wt MS7163 as determined by qRT-PCR. Total mRNA was extracted from bacteria grown on YESCA agar at 37°C for 24 h. Results are displayed as the mean log2 FC with standard deviation from three independent replicates. (C) CR staining of wt MS7163, MS7163csgA, Tn5-03831, PcL-rcpA, MS7163(pRcpA), and MS7163(pSU2718 [vector control]) following growth on YESCA-CR agar. Strains were grown by spotting 5 µl of an overnight culture on YESCA-CR agar and incubating for 24 h at 37°C. (D) Western blot analysis of CsgA performed using whole-cell lysates prepared from MS7163, MS7163csgA, Tn5-03831, PcL-rcpA, MS7163(pRcpA), and MS7163(pSU2718). Bacteria were grown on YESCA agar for 24 h to induce curli production and treated with formic acid to dissolve polymerized CsgA. Anti-OmpA antibody was used as a loading control.