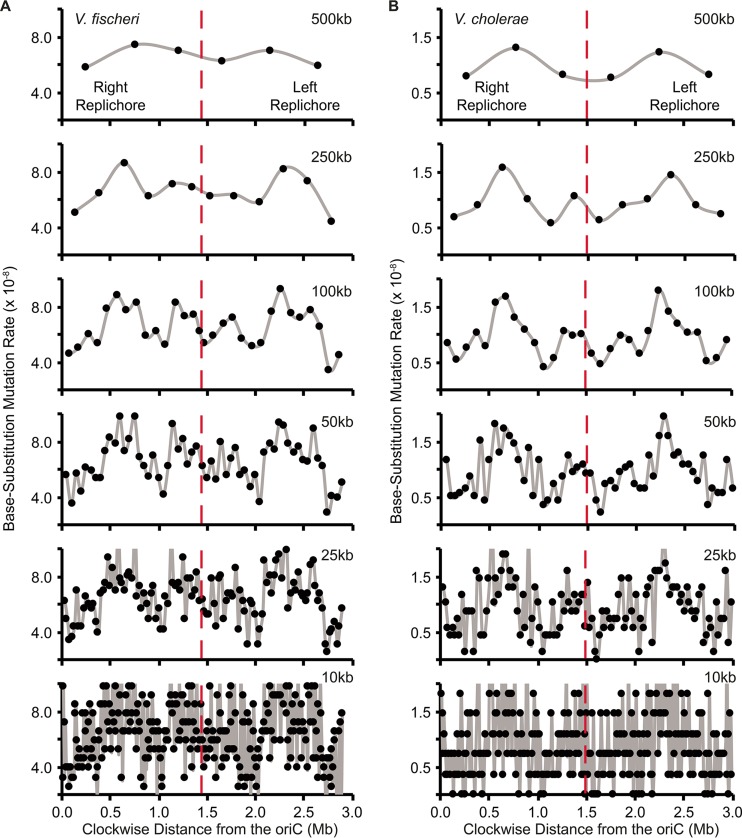
FIG 1 .
Patterns of base substitution mutation (bpsm) rates at various size intervals extending clockwise from the origin of replication (oriC) in MMR-deficient mutation accumulation lineages of V. fischeri (A) and V. cholerae (B) on chromosome 1. bpsm rates are calculated as the number of mutations observed within each interval divided by the product of the total number of sites analyzed within that interval across all lines and the number of generations of mutation accumulation. The two intervals that meet at the terminus of replication (dotted red line) on each replichore are shorter than the interval length for that analysis, because the size of chromosome 1 is never exactly divisible by the interval length.