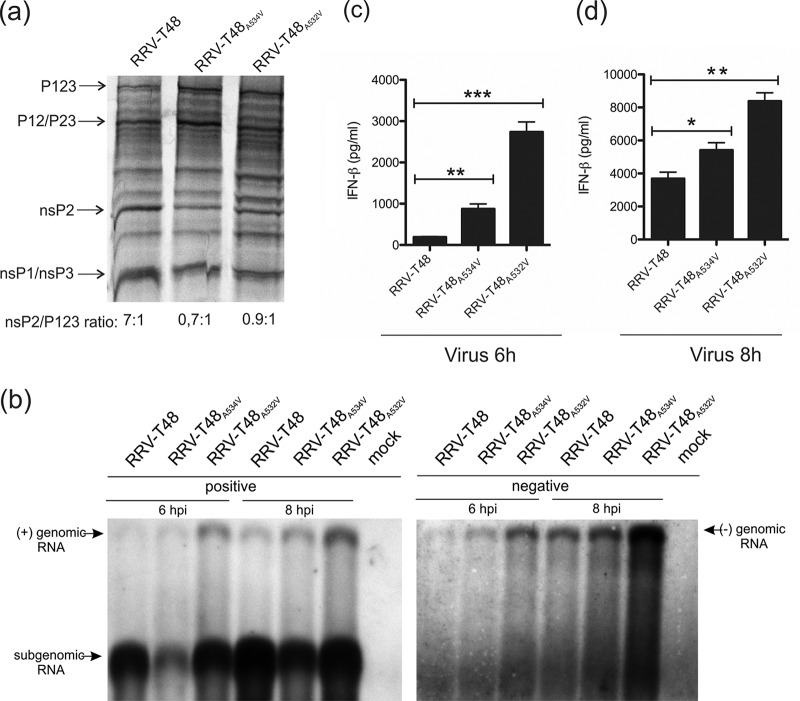
FIG 4 .
Effects of mutations in the P3 and P1 positions of the 1/2 site on RRV nonstructural polyprotein processing and RNA replication. (a) Processing of RRV-T48, RRV-T48A532V and RRV-T48A534V nonstructural polyproteins was analyzed as described in the legend to Fig. 3a. (b to d) BHK-21 cells were infected with RRV-T48, RRV-T48A532V or RRV-T48A534V at an MOI of 10. At 6 or 8 h p.i., cells were lysed, and total RNA was isolated. (b) Northern blot analysis. RNAs isolated from infected cells were separated by electrophoresis in agarose gels, transferred to a Hybond-N+ filter, and detected with probes complementary to positive and negative strands of RRV RNAs. (c and d) One microgram of each RNA sample was used for transfection of Cop5 cells; at 24 h p.t., the amount of IFN-β in the cell supernatant was determined. IFN-β amounts are expressed as the means ± SEM from three experiments performed in parallel (*, P < 0.05; **, P < 0.01; ***, P < 0.001 using Student’s two-tailed unpaired t test). Data from one of two reproducible independent experiments are shown in each of the panels.