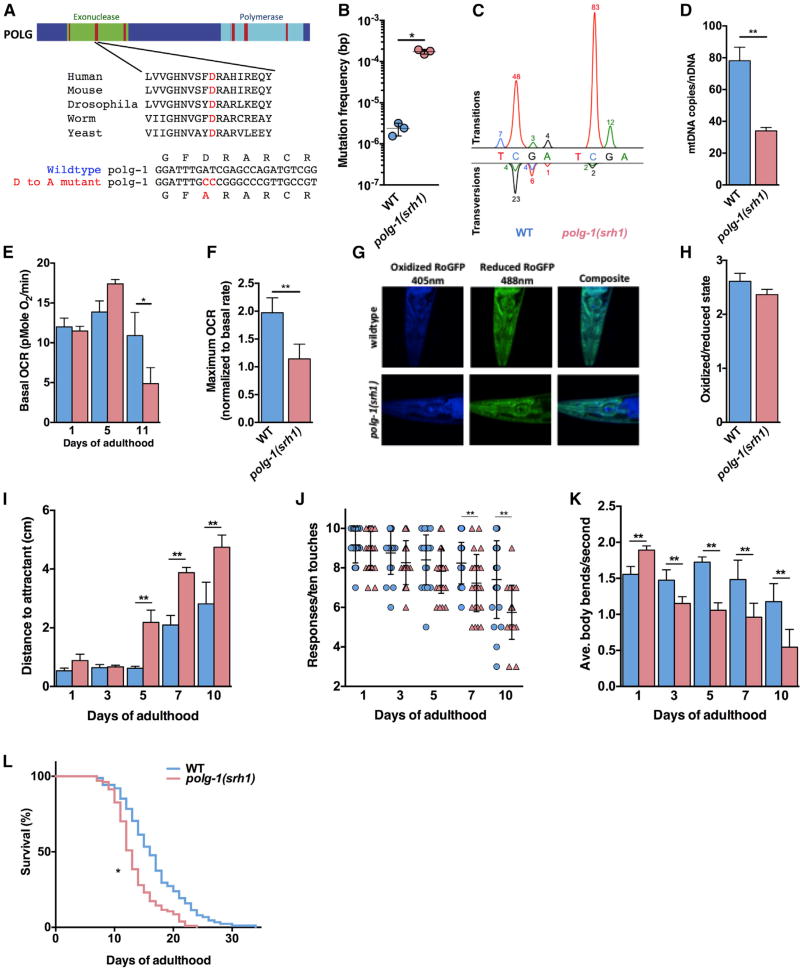
Figure 1. Characterization of the polg-1(srh1) Worms.
(A) The exonuclease (green) domain of DNA polymerase γ contains three highly conserved regions (red) that control the fidelity of DNA synthesis, including an aspartic acid in exonuclease domain II that is essential for the proofreading activity of polymerase γ across the tree of life. We used CRISPR/Cas9 technology to mutate this residue to alanine in the error-prone allele polg-1(srh1).
(B) The mtDNA mutation frequency of the polg-1(srh1) worms is >70-fold higher compared to WT worms.
(C) The mutation spectrum of WT and polg-1(srh1) depicted as peaks above and below the WT nucleotide according to standard electrophoretogram color coding (red, T; blue, C; black, G; green, A). The percentage of each type of mutation is listed by the peaks.
(D) The mtDNA copy number is reduced by 56% in the polg-1(srh1) worms.
(E) The basal respiration of polg-1(srh1) worms worsens with age compared to WT worms.
(F) The mitochondria of polg-1(srh1) worms display reduced reserve capacity upon FCCP treatment at 5 days of age.
(G and H) Confocal images of day 10 polg-1(srh1) and WT worms (G) do not reveal a significant difference in oxidation stress (H).
(I–K) Neuromuscular function assessed by chemotaxis (I), thrashing (J), and a gentle touch assay (K) reveals increased dysfunction in polg-1(srh1) worms compared to WT animals.
(L) The polg-1(srh1) worms have a median lifespan of 13 days compared to 16 days for WT worms (log-rank test p < 0.01).
Data for WT and polg-1(srh1) worms are in blue and pink, respectively (in B, D–F, and H–L).
Bar graphs represent the mean ± SEM of at least three biological replicates, and the lifespan assay was performed using at least 100 worms per genotype. Unpaired t tests were performed to determine significance (*p < 0.05, **p < 0.01; ns, no significant difference).