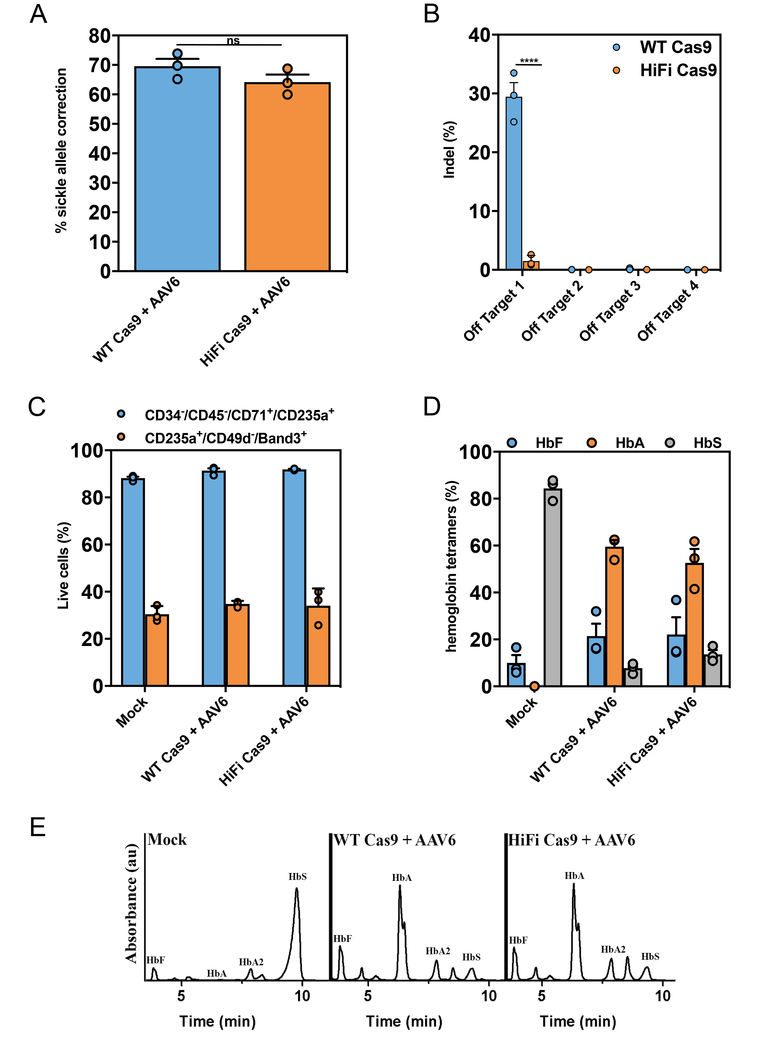
Figure 6. R691A HiFi Cas9 delivered as RNP mediates robust Glu6Val HBB gene correction in sickle cell disease-CD34+ HSPCs while significantly reducing off-target activity.
(a) Sickle cell disease (SCD) patient-derived HSPCs were isolated from peripheral blood and subjected to Glu6Val gene correction methods using HBB RNPs (either WT (blue) or HiFi (orange) Cas9) and a gene corrective ssAAV6. Gene correction frequencies were analyzed by ddPCR™ as described in materials and methods (Bars represent mean ± s.e.m., n =3, number of data points within each group, all from different SCD patients). NS (not significant) = P ≥ 0.05, Two-tailed Student’s T-test. (b) SCD-HSPCs were targeted for Glu6Val HBB gene correction as described above and off-target editing was analyzed by NGS. Bars represent mean ± s.e.m., n=3 SCD patients. ****P < 0.0001, NS (not significant) = P ≥ 0.05, two-way analysis of variance (ANOVA) and Tukey’s multiple comparisons test. (c) Glu6Val targeted WT or HiFi SCD-HSPCs were differentiated down the erythroid lineage ex vivo. At 14 days post-differentiation, cells were harvested and evaluated for the percentage of erythrocytes (CD34−/CD45−/CD71+/CD235a+) and reticulocytes (CD235a+/CD49d−/Band3+) by FACS-based immunophenotypic analyses. Bars represent mean ± s.e.m., n=3 SCD patients. (d) Glu6Val targeted erythrocyte differentiated HSPCs were subjected to cation-exchange HPLC analysis of steady-state hemoglobin tetramers. The normalized percentages of fetal (HbF), adult (HbA) and sickle (HbS) hemoglobin are plotted as a function of total hemoglobin tetramers on the HPLC plot (as presented in Fig. 6f). Hemoglobin tetramers were quantified by measuring the area under the curve (AUC) of the absorbance peaks. Bars represent mean ± s.e.m., n=3 SCD patients. (e) Representative HPLC plot from the data presented in Fig. 6d showing Glu6Val correction in SCD-HSPCs results in HbA protein production when using both the WT and HiFi Cas9 HBB RNPs. Experiment is representative of the 3 experiments performed in Fig. 6d with similar results.