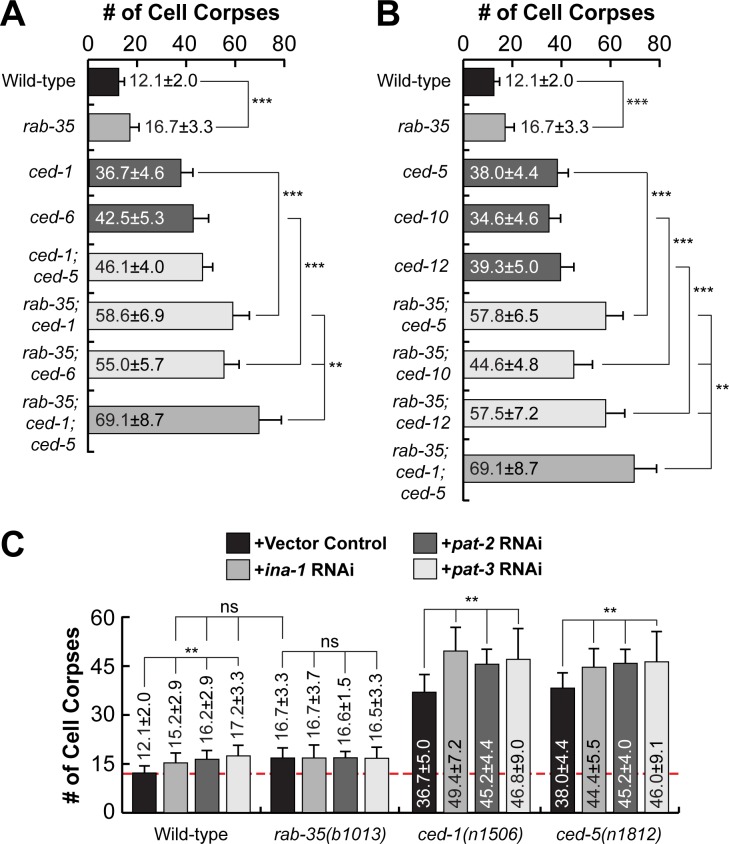
Fig 10. rab-35 in an engulfment pathway independent of both the ced-1 and ced-5 pathways yet includes the integrins.
Results of epistasis analysis performed between rab-35 and (A) members of the ced-1/-6/-7 pathway, (B) members of the ced-2/-5/-10/-12 pathway, and (C) genes encoding C. elegans integrins. The average numbers of cell corpses observed in 1.5-stage embryos of various genotypes are listed and presented as bars. Error bars indicate sd. For each strain, at least 15 animals were scored. Student t-test was used for data analysis: *, 0.001 < p < 0.05; **, 0.00001 < p <0.001; ***, p <0.00001; ns, no significant difference. (A) Null alleles [rab-35(b1013), ced-1(n1506), and ced-6(n2095)] were used. (B) Null alleles [ced-5(n1812) and ced-12(n3261)] and a severe loss-of-function allele ced-10(n1993) were used. (C) C. elegans integrin genes ina-1, pat-2, and cdc-42 were inactivated in wild-type, rab-35(b1013), ced-1(n1506), and ced-5(n1812) backgrounds by RNAi. A dashed line represents the mean number of cell corpses observed in untreated wild-type embryos.