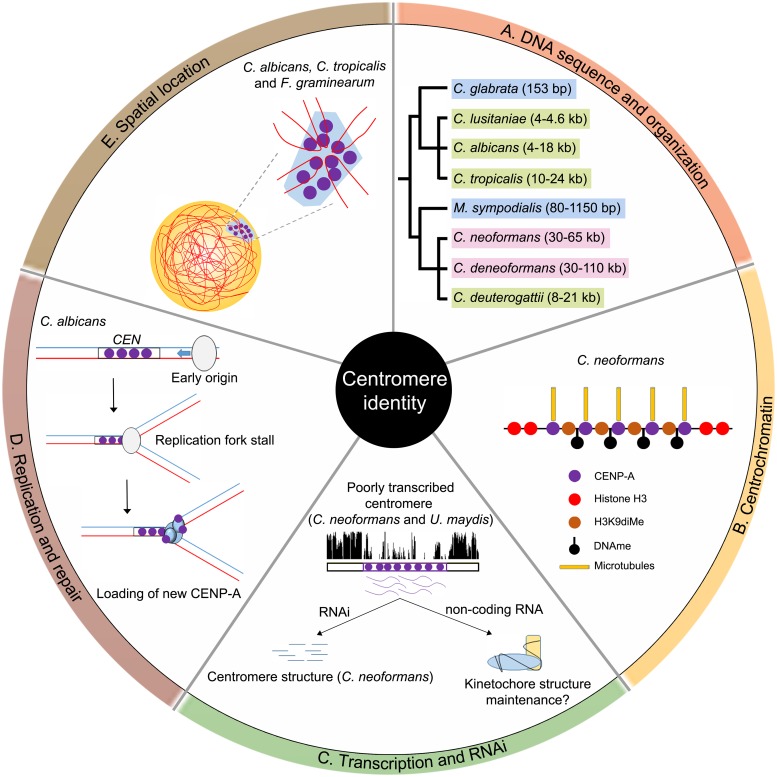
Fig 1. Five key determinants of centromere identity in pathogenic fungi.
(A) The length and type of centromeres in various pathogenic fungi have been depicted in a cladogram. Candida glabrata and Malassezia sympodialis have point centromeres (blue); C. albicans, C. tropicalis, and Cryptococcus deuterogattii have small regional centromeres (green); and C. neoformans and C. deneoformans harbor large regional repetitive centromeres (pink). (B) Centromeric heterochromatin or centrochromatin refers to the various chemical modifications associated with the histones (both canonical and variant) as well as DNA at the centromeric locus. For example, centromere DNA is methylated at cytosines, and CENP-A nucleosomes are interspersed with H3K9-dimethylated nucleosomes in C. neoformans. (C) Transcription and a functional RNAi machinery at the centromere are required to preserve centromere identity in some organisms. The centromeres in the Ustilago and Cryptococcus species complexes are poorly transcribed. In C. neoformans, centromeric transcription is regulated by the RNAi machinery that silences centromeric retrotransposons to stabilize centromere structure. Unprocessed long noncoding RNA, whose function is unknown, is also produced from centromeres in this species. (D) DNA replication and repair proteins are known to play a key role in C. albicans centromere stability. The centromere proximal origins help in maintaining centromere function in this organism. Replication forks converging toward the centromere stall randomly, which leads to accumulation of single-stranded DNA (ssDNA) that recruits repair proteins like Rad51 and Rad52, along with new CENP-A molecules in C. albicans. (E) The spatial location of centromeres within the nucleus determines its activity and interaction with other nuclear subcompartments. All centromeres are clustered toward the nuclear periphery to form a CENP-A–rich zone in C. albicans and many other pathogenic fungi. This preferential spatial distribution helps to determine the site of centromere assembly in every cell cycle. CENP-A, centromere protein A; H3K9, histone H3 lysine 9; Rad, radiation sensitive; RNAi, RNA interference; ssDNA, single-stranded DNA.