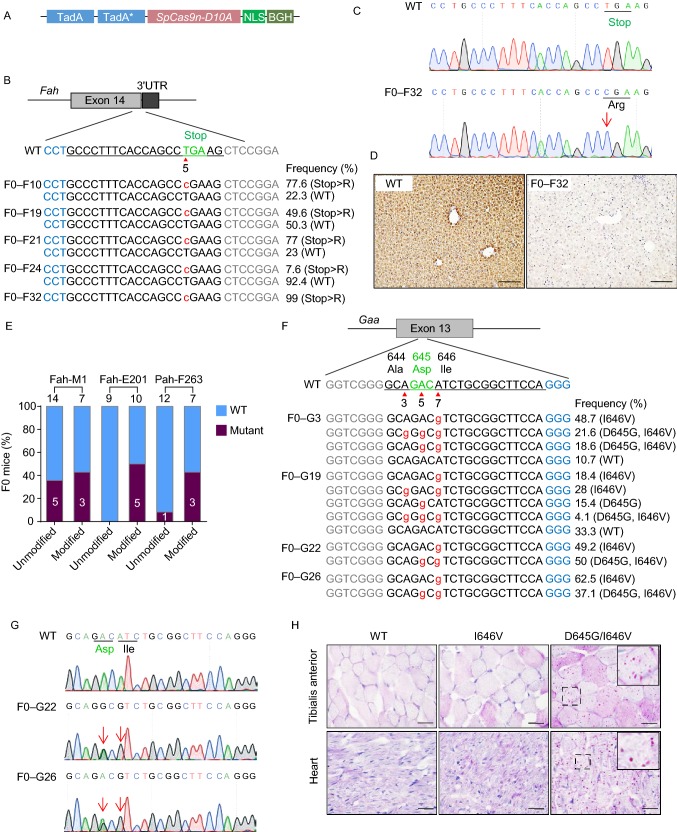
Figure 1.
ABE induces efficient A>G conversion in mouse and rat embryos. (A) A schematic view of the ABE7.10 vector used as the template for ABE mRNA transcription. (B) A schematic view of the target site at the Fah stop codon. Target sequence is underlined. PAM sequence is labeled in blue. Stop codon is labeled in green. Arrow head indicates the targeted thymine. Base substitutions are labeled in red. Allele frequencies are listed to the right. (C) Sanger sequencing chromatograms from the WT and F0–F32 founder. T>C conversion is indicated by the red arrow. (D) IHC staining of the liver tissue sections from WT and F0–F32 founder by anti-Fah antibody. Scale bar, 100 μm. (E) The editing efficiencies at three different target sites with chemically modified crRNA/tracrRNAs or unmodified sgRNAs. The numbers indicate the number of pups generated. (F) A schematic view of the target site in exon 13 of the rat Gaa gene and deep sequencing results from the genomic DNA of the mutant founders. PAM sequence is labeled in blue. Target sequence is underlined with codon 644, 645 and 646 indicated by their amino acid. Base substitutions are labeled in red. Allele frequencies are listed to the right. (G) Sanger sequencing chromatograms from the genomic DNA of WT and two mutant F0 founders. Double peak signals caused by A>G conversions are indicated by red arrows. Codon 645 and 646 of WT and mutant alleles are underlined. (H) PAS staining of heart and Tibialis anterior cryo-sections from 3 week old WT, I646V and D645G/I646V homozygotes. Scale bar, 20 μm