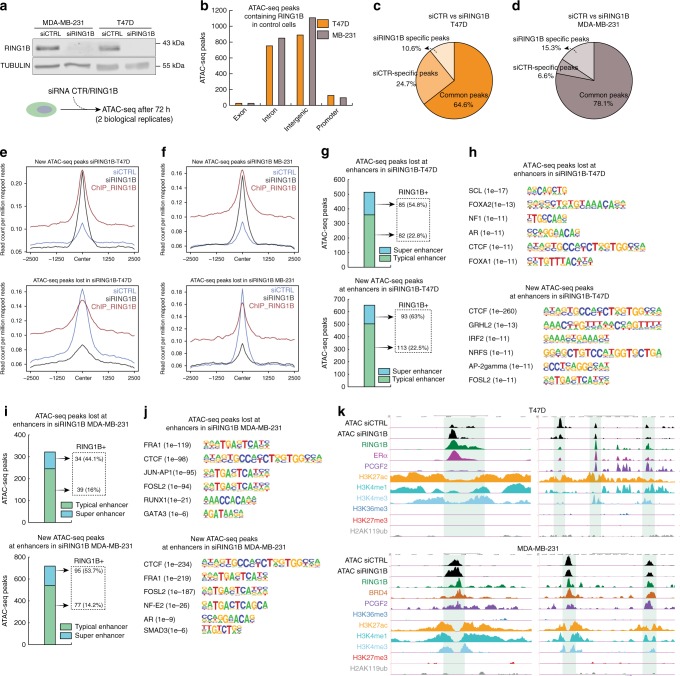
Fig. 6.
RING1B regulates chromatin accessibility at enhancers. a Western blot of RING1B from control and RING1B-depleted cells 72 h after siRNA transfection. TUBULIN was used a loading control. ATAC-seq experiments were performed in two biological replicates after siRNAs transfections. b ATAC-seq peak distribution in genomic sites bound by RING1B in RING1B-depleted cells. c, d Pie charts showing percentage of ATAC-seq peaks not affected by RING1B depletion (common peaks), lost after RING1B depletion (siCTR-specific peaks), or gained after RING1B depletion (siRING1B-specific peaks. Two ATAC-seq experiments were performed after two independent siRING1B transfections. e, f RING1B ChIP-seq signals and ATAC-seq signals at acquired and lost ATAC-seq peaks. g ATAC-seq peaks at enhancers after RING1B depletion, number of enhancers containing RING1B ChIP-seq signals in T47D. h Transcription factor-binding motif analysis of peaks acquired or lost at enhancers in T47D. i Acquired and lost ATAC-seq peaks at enhancers after RING1B depletion, number of enhancers containing RING1B ChIP-seq signals in MDA-MB-231. j Transcription factor-binding motif analysis in peaks acquired or lost at enhancers in MDA-MB-231. k Genome browser screenshots of ChIP-seq and ATAC-seq profiles at selected enhancers