Abstract
Background: Crohn's disease and Ulcerative colitis are known as inflammatory bowel disease with high morbidity which are as a result of increasing immune responses to intestinal microbiota in genetically susceptible individuals. The association of adherent invasive Escherichia coli with Crohn's disease in human has been discussed for decades. The principal aim of this study was to assess the relationship between adherent invasive Escherichia coli in Iranian patients with Crohn's disease.
Methods: The presence of adherent invasive Escherichia coli DNA and viable adherent invasive Escherichia coli cells were identified through PCR and conventional culture methods, respectively. All the specimens were subsequently cultured in Hi Chrome Agar medium.
Results: Using molecular assay, the invasive plasmid antigen H and invasion-association locus genes were detected from tissue samples confirming the presence of adherent-invasive Escherichia coli. The invasive plasmid antigen H was detected in 46.7% of CD and 13.3% of healthy peoples. The invasion-association locus gene was found in 36.7% of patients with Crohn's disease and 10% in individuals without IBD.
Conclusion: This study demonstrated an increased frequency of adherent invasive E. coli with invasive plasmid antigen H and invasion-association locus genes from patients with CD in comparison to control individuals. Moreover, it was shown that adherent invasive E. coli with the invasive plasmid antigen H and invasion-association locus genes can act as a predisposing factor in the development of IBD.
Keywords: Crohn Disease, Inflammatory bowel disease, Escherichia coli, PCR
↑ What is “already known” in this topic:
Inflammatory bowel disease (IBD) is comprised of Crohn's disease (CD) and Ulcerative colitis (UC). The prevalence and incidence of IBD are continuously rising in Iran. No study has been conducted to investigate the role of adherent invasive E. coli in patients with Crohn's disease occurred in the country.
→ What this article adds:
Due to the rising trend of IBD prevalence and incidence in Iran, and also the significant role of intestinal microbiota in the development of disease, further investigation into the possible causing factors is vitally imperative.
Introduction
Inflammatory bowel disease (IBD) is a chronic inflammatory disorder divided into Ulcerative colitis (UC) and Crohn’s disease (CD) (1, 2).The rising trend of the disease has observed in Middle Eastern countries as well as in other parts of the world and statistics shows the incidence also is on the rises in North America and Europe as well as other parts of the world. Likewise, a recent study demonstrated that the prevalence of IBD is increasing dramatically in Iran (3).
The incidence and prevalence of ulcerative colitis and Crohn's disease have been increased all over the world, especially among children and adults. It is believed that several factors such as immunology, genetic factors, stress, diet, being an alcoholic or a smoker, as well as bacteria such as the adherent-invasive strains of Escherichia coli, Campylobacter spp., Clostridium dificil, Mycobacterium avium subspecies Paratuberculosis, and Bacteroides fragilis (4-7) may play a role in the development of IBD. Since gastrointestinal (GI) tract of healthy people is colonized by a wide variety of different microorganisms hence, dysbiosis of the intestinal microbiota is likely cause of IBD (8).
It is known that intestinal bacteria can cause CEACAM6 expression through inducing inflammatory cytokines. The expression can then leads to production of E. coli with AIEC phenotype. The AIEC can attach to epithelial cells by a mechanism involving polymerization of microtubules. In fact, AIEC adheres to GP2 (glycoprotein 2) upon M cells by LPF (long polar fimbriae) and enter to Peyer's patches and exacerbates inflammation in the intestine (9). Moreover, AIEC able to adhere epithelial cells and invade into the cytoplasmic eukaryotic infectious cells with type 1fimbriae (fimH), invasive plasmid antigen H (ipaH) and invasion-association locus (ial) (10).
However, E. coli with adherent-invasive phenotype should be regarded as a separate issue of pathogenicity of E. coli causing inflammation in the human intestine.
The principal purpose of this study was to investigate of E. coli with adherent-invasive phenotype and their relation to Crohn’s disease.
Methods
In the present study, sixty subjects including thirty patients with lesions of Crohn's colitis and thirty people without IBD were collected during colonoscopy examinations. Samples with CD were obtained from lesions of colon, or terminal ileum. Moreover, the biopsy of control group was performed on normal areas. DNA was extracted from the tissue samples and PCR assay performed by targeting ipaH and ial genes. CD diagnosis based on clinical symptoms, laboratory evaluations, and colonoscopy finding was confirmed by histological assessment. The control subjects were taken from people with non-inflammatory IBD (nIBD). None of the individuals presented in this study used antibiotics or probiotics for the 3 months prior to the study. The study was accepted by the ethics committee of Tehran University Medical Sciences.
Microbial identification
Samples were immediately transferred into sterile vials containing either thioglycolate broth or saline (Sigma-Aldrich, Hi Media) and stored at -20˚C. The tissue biopsy were homogenized and inoculated into Hi Chrome E. coli agar (Sigma-Aldrich, Hi Media) and incubated for 18-24h at 35±2°C. The bacteria were stored in TSB broth containing 30% glycerol at a temperature of -70°C for further analysis.
DNA Extraction
Tissue biopsies were crushed and DNA extracted through RTP® Mycobacteria kit (Berlin, Germany).
PCR assay
All samples were tested for the presence of ipaH and ial genes using molecular PCR. The nucleotide sequence of primers (Macrogen, Pishgam) and production size (base pairs) for amplification of the ipaH and ial genes are shown in Table 1 (11, 12). PCR was performed in 12.5μL comprising 5μL master mix (Amplicon, Pishgam), 0.5 μL of each primer, 2 μL of the DNA template (50 ng) and 4.5μL of ddH2O. Subsequently, the thermal cycling status was used: 300 seconds at 94°C and 25 cycles of amplification consisting of 60 seconds at 95°C, 55 seconds at 58°C, and 60 seconds at 72°C, with 300 seconds at 72°C for the final extension. PCR products were investigated by electrophoresis on a 1% agarose gel in 1X TBE buffer [10.8 g Tris and 5.5 g Boric acid, 0.5 M Na2EDTA (pH 8.0)] (13).
Table 1. PCR Analysis of the ipaH gene for CD and nIBD samples .
| Genes | Nucleotide Sequences(5´-3´) | Size of product, bp |
| ipaH |
F-GTTCCTTGACCGCCTTTCCGATACCGTC R-GCCGGTCAGCCACCCTCTGAGAGTAC |
619 |
| ial |
F-CTGGATGGTATGGTGAGG R-GGAGGCCAACAATTATTTCC |
320 |
Statistical Analysis
Data were analyzed using Pearson Chi-Square test, and P value below 0.05 considered as statistically significant.
Results
Clinical information of patients and controls
The average age of patients with IBD and controls were 38.8 and 52.86 years old respectively. The participants included 14 males and 16 female patients with CD, and 18 males and 12 females without IBD. In addition, the age range for patients with CD and without IBD was 18-64 and 25-76 years old respectively.
Detection of adherent-invasive E. coli with ipaH and ial genes
Of 60 tissue samples collected, E. coli were isolated from 20 (66.6%) patients with CD and 22 (73.3%) from healthy people (Table 2). All isolated bacteria were confirmed as AIEC by biochemical tests and PCR assay. The ipaH, ial genes were amplified by utilizing particular primers and become visible as a band of approximately 619 and 320 kb on agarose gel respectively (Figs. 1 and 2). The difference between patients with Crohn’s disease and controls was not significant (p=0.59). Of 30 patients with CD, the positive and negative rate of ipaH gene was 46.7% (n=14) and 53.3% (n=16) respectively.
Table 2. E. coli Bacterial Culture in various condition .
| Group | Culture of | Total | ||
| Positive | Negative | |||
|
Crohn’s disease Count |
10 | 20 | 30 | |
| CD (%) | 33.4 | 66.6 | 100.00 | |
|
nIBD
Count |
8 | 22 | ||
| nIBD (%) | 26.7 | 73.3 | 100.00 | |
| Total Count | 18 | 42 | 60 | |
| Total (%) | 30 | 70 | 100.00 | |
Fig. 1.
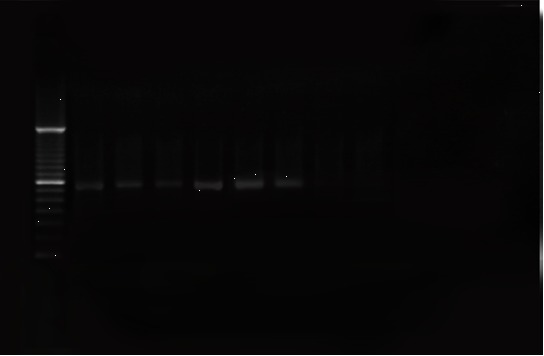
Amplification of the ipaH gene. Lanes: 1, DNA molecular size marker (100–bp ladder); 2-7, Crohn’s disease samples; 8, negative control (Water).
Fig. 2.
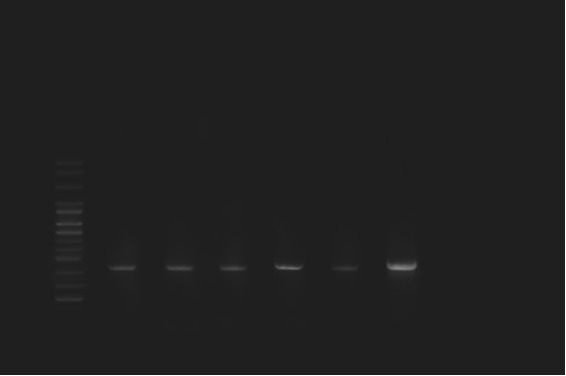
Amplification of the ial gene. Lanes: 1, DNA molecular size marker (100–bp ladder); 2-7, Crohn’s disease samples; 8, negative control (Water).
The percentage of ipaH among 30 individuals without IBD was 13.3% (n=4) but 86.6% (n=26) was not detected (Table 3). Furthermore, the positivity of ial gene were 36.7% (n=11) and 10% (n=3) in patients with CD and healthy people respectively (Table 4). Moreover, the presence of ipaH and ial genes in adherent-invasive E. coli in patients with CD was more than control populations.
Table 3. PCR Analysis of the ipaH gene for CD and nIBD samples .
| Group | ipaH | |
| Positive | Negative | |
| Crohn’s disease Count | 14 | 16 |
| CD (%) | 46.7 | 53.3 |
| P value nIBD | .005* | |
| Count | 4 | 26 |
| nIBD (%) | 13.3 | 86.7 |
| P value | 0.24 | |
*Significant at alpha=0.05
Table 4. PCR Analysis of the ial gene from CD and nIBD samples.
| Group | ial | |
| Positive | Negative | |
| Crohn’s disease | ||
| Count | 11 | 19 |
| CD (%) | 36.7 | 63.3 |
| P value | .015* | |
| nIBD | ||
| Count | 3 | 27 |
| nIBD (%) | 10.0 | 90.0 |
| P value | 0.34 | |
Due to its high sensitivity, PCR assay was more reliable than cultivation. The difference between the correlation of adherent-invasive E. coli with ipaH and ial in CD patients with control populations was statistically significant (p<0.05). All positive amplified fragments were sequenced and high percentage of adherent-invasive E. coli was identified.
Furthermore, a chi square test was performed and the correlation between adherent-invasive E. coli with ipaH and ial genes in patients with CD found to be (N=30) =7.937, p= .005 and (N=30) =5.963, p=.015 respectively. Nonetheless all positive amplified fragments were sequenced and high percentage of adherent-invasive E. coli was detected.
Discussion
Crohn's disease is considered as a chronic inflammatory of GI disorder.
Although the etiology of inflammatory bowel disease is not still known (14), but numerous studies demonstrated that the mortality rate of this disease is high (15). Furthermore, it is demonstrated that several factors may play role in induction of the disease. In a research by Sartor RB et al, the intestinal microbiota effect on epithelial cells, and inflammation of the intestinal mucosa have shown to trigger the pathogenesis of both CD and UC (16). Moreover, CD patients demonstrated an altered intestinal microbial community, with different kind of bacteria in colonic and ileal areas (17).
Previous studies showed that Enterobacteriaceae especially E. coli could contributed the onset of IBD. Nevertheless other bacteria can trigger the pathogenesis of inflammatory bowel disease including Bacteroidales, Clostridiales, Pasteurellaceae (Haemophilus sp.), Veillonellaceae, Neisseriaceae, and Fusobacteriaceae (18). Significant information exists regarding the role of adherent invasive E. coli in the CD. Numerous studies reported that adherent invasive E. coli could be a candidate for the development of CD (19).
An emerging evidence indicated an association between adherent invasive E. coli with IBD and colorectal cancer (20, 21 and 22). The present study shows no difference in the incidence of E. coli positive culture between CD and control patients (Table 2). The lack of E. coli in some tissue sample was probably due to consumption of antibiotics which in turn can cause dysbiosis in the human gastrointestinal tract (23). In some studies, using the culture-based method demonstrated that E. coli bacteria were found in the intestinal mucosa as well as in ulcers of both CD and UC patients compared to controls (24). In comparison with nIBD, our finding shows that the abundance of adherent invasive E. coli harboring ipaH and ial genes was higher in CD individuals. Additionally, we found that 46.7% and 36.7% of CD specimens harbored ipaH and ial, an increased proportion of AIEC, compared to control individuals. In spite of interesting debate, Dogan et al conducted a similar prevalence of AIEC in the ileum of patients with CD (25). Several studies showed a high proportion of AIEC harboring fimH, fimA and Cas genes in CD (26, 27). According to the present study, in comparison with nIBD, PCR assay displayed a high abundance of ipaH and ial genes in CD patients with statistically significant difference. These data proposed that AIEC most likely harbored ipaH and ial genes which in turn could be effective in the initiation of the CD. Nonetheless, since other virulence factors for AIEC were not analyzed, so it may be taken into consideration in the future study.
Conclusion
Despite the role of several factors in the progression of IBD, the present study displays a high prevalence of AIEC with ipaH and ial from patients with CD. According to these findings, E. coli with AIEC phenotype could be a predisposing factor in the development of inflammatory bowel disease. In addition to AIEC, host immune response, genetics of patients and environmental factors play an important role in the initiation of IBD. Therefore the present study suggests that appropriate treatment against AIEC could be practically effective in patients with inflammatory bowel disease.
Acknowledgments
We would like to thank the Behbood clinical staff for their kind cooperation. This study is supported by the Tehran University of Medical Sciences.
Funding/Support
This study was supported in part by grant No. 33309 provided by Tehran University of Medical Sciences.
Conflict of interests
The authors declare that they have no competing interests.
Cite this article as: Sarabi Asiabar A, Asadzadeh Aghdaei H, Sabokbar A, Zali MR, Feizabadi MM. Investigation of adherent-invasive E. coli in patients with Crohn's disease. Med J Islam Repub Iran. 2018(11 Feb);32:11. https://doi.org/10.14196/mjiri.32.11
References
- 1.Balfour Sartor MDR Balfour Sartor MDR, Mazmanian SK Mazmanian SK. Intestinal Microbes in Inflammatory Bowel Diseases. Am J Gastroenterol Suppl. 2012 Jul;1(1):15. [Google Scholar]
- 2.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci. 2008;105(52):20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taghavi SA, Safarpour AR, Hosseini SV, Noroozi H, Safarpour M, Rahimikazerooni S. Epidemiology of inflammatory bowel diseases (IBD) in Iran: a review of 740 patients in Fars Province, Southern Iran. Ann Colorectal Res. 2013;1(1):17–22. [Google Scholar]
- 4.Ng SC, Tang W, Leong RW, Chen M, Ko Y, Studd C. et al. Environmental risk factors in inflammatory bowel disease: a population-based case-control study in Asia-Pacific. Gut . 2014 doi: 10.1136/gutjnl-2014-307410. [DOI] [PubMed] [Google Scholar]
- 5.Hold GL, Smith M, Grange C, Watt ER, El-Omar EM. Role of the gut microbiota in inflammatory bowel disease pathogenesis: What have we learnt in the past 10 years? World J Gasterolentrol. 2014;20(5):1192–1210. doi: 10.3748/wjg.v20.i5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmed I, Roy BC, Khan SA, Septer S, Umar S. Microbiome, metabolome and inflammatory bowel disease. Microorganisms. 2016 Jun 15;4(2):20. doi: 10.3390/microorganisms4020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sartor RB. Genetics and environmental interactions shape the intestinal microbiome to promote inflammatory bowel disease versus mucosal homeostasis. Gastroenterology. 2010;139:1816–1819. doi: 10.1053/j.gastro.2010.10.036. [DOI] [PubMed] [Google Scholar]
- 8.Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P. et al. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;1:2010. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 9.Martinez-Medina M, Garcia-Gil LJ. Escherichia coli in chronic inflammatory bowel diseases: an update on adherent invasive Escherichia coli pathogenicity. World J Gastrointest Pathophysiol. 2014;5(3):213. doi: 10.4291/wjgp.v5.i3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chassaing B, Rolhion N, de Vallée A, Sa’ad YS, Prorok-Hamon M, Neut C. et al. Crohn disease–associated adherent-invasive E coli bacteria target mouse and human Peyer’s patches via long polar fimbriae. J Clin Invest. 2011;121(3):966–75. doi: 10.1172/JCI44632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Souza HL, de Carvalho VR, Romeiro FG, Sassaki LY, Keller R, Rodrigues J. Mucosa-associated but not luminal Escherichia coli is augmented in Crohn’s disease and ulcerative colitis. Gut Pathog. 2012;4(1):21. doi: 10.1186/1757-4749-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cruz CB, Souza MC, Serra PT, Santos I, Balieiro A, Pieri FA, et al. Virulence factors associated with pediatric shigellosis in Brazilian Amazon. Biomed Res Int 2014; 2014. [DOI] [PMC free article] [PubMed]
- 13.Zhang Y, Rowehl L, Krumsiek JM, Orner EP, Shaikh N, Tarr PI. et al. Identification of candidate adherent-invasive E coli signature transcripts by genomic/transcriptomic analysis. PloS One. 2015;10:e0130902. doi: 10.1371/journal.pone.0130902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez SM, Martinez MM, Busquets D. “Mucosa-associated Faecalibacterium prausnitzii and Escherichia coli co-abundance can distinguish Irritable Bowel Syndrome and Inflammatory Bowel Disease phenotypes”. Int J Med Microbiol. 2014;304(4):464–475. doi: 10.1016/j.ijmm.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Bitton A, Vutcovici M, Sewitch M, Suissa S, Brassard P. Mortality trends in Crohn's disease and ulcerative colitis: a population-based study in Quebec, Canada. Inflamm Bowel Dis. 2016;22(2):416–23. doi: 10.1097/MIB.0000000000000608. [DOI] [PubMed] [Google Scholar]
- 16. Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134(2):577–94. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 17.Sheehan D, Moran C, Shanahan F. The microbiota in inflammatory bowel disease. J Gastroenterol. 2015;50(5):495–507. doi: 10.1007/s00535-015-1064-1. [DOI] [PubMed] [Google Scholar]
- 18.Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R. et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 19.Strober W. Strober WAdherent-invasive Ecoli in Crohn disease: bacterial “agent provocateur”J Clin Invest. 2011 Mar. 1;121(3):841. doi: 10.1172/JCI46333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X. et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6(2):320–9. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iebba V, Conte MP, Lepanto MS, Di Nardo G, Santangelo F, Aloi M. et al. Microevolution in fimH gene of mucosa-associated Escherichia coli strains isolated from pediatric patients with inflammatory bowel disease. Infect Immun. 2012;80(4):1408–17. doi: 10.1128/IAI.06181-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B. et al. Colonization of the human gut by E coli and colorectal cancer risk. Clin Cancer Res. 2014;20(4):859–67. doi: 10.1158/1078-0432.CCR-13-1343. [DOI] [PubMed] [Google Scholar]
- 23.Small CL, Reid-Yu SA, McPhee JB, Coombes BK. Persistent infection with Crohn's disease-associated adherent-invasive Escherichia coli leads to chronic inflammation and intestinal fibrosis. Nat Commun. 2013;4:1957. doi: 10.1038/ncomms2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conte MP, Longhi C, Marazzato M, Conte AL, Aleandri M, Lepanto MS. et al. Adherent-invasive Escherichia coli (AIEC) in pediatric Crohn’s disease patients: phenotypic and genetic pathogenic features. BMC Res Notes. 2014;7(1):748. doi: 10.1186/1756-0500-7-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dogan B, Scherl E, Bosworth B, Yantiss R, Altier C, McDonough PL. et al. Multidrug resistance is common in Escherichia coli associated with ileal Crohn's disease. Inflamm Bowel Dis. 2013;19(1):141–50. doi: 10.1002/ibd.22971. [DOI] [PubMed] [Google Scholar]
- 26.Gombošová L, Lazúrová I, Zakuciová M, Čurová K, Kmeťová M, Petrášová D. et al. Genes of intestinal Escherichia coli and their relation to the inflammatory activity in patients with ulcerative colitis and Crohn’s disease. Folia Microbiol. 2011 Sep 1;56(5):367. doi: 10.1007/s12223-011-0051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Rowehl L, Krumsiek JM, Orner EP, Shaikh N, Tarr PI. et al. Identification of candidate adherent-invasive E coli signature transcripts by genomic/transcriptomic analysis. PloS One. 2015;10(6):e0130902. doi: 10.1371/journal.pone.0130902. [DOI] [PMC free article] [PubMed] [Google Scholar]


