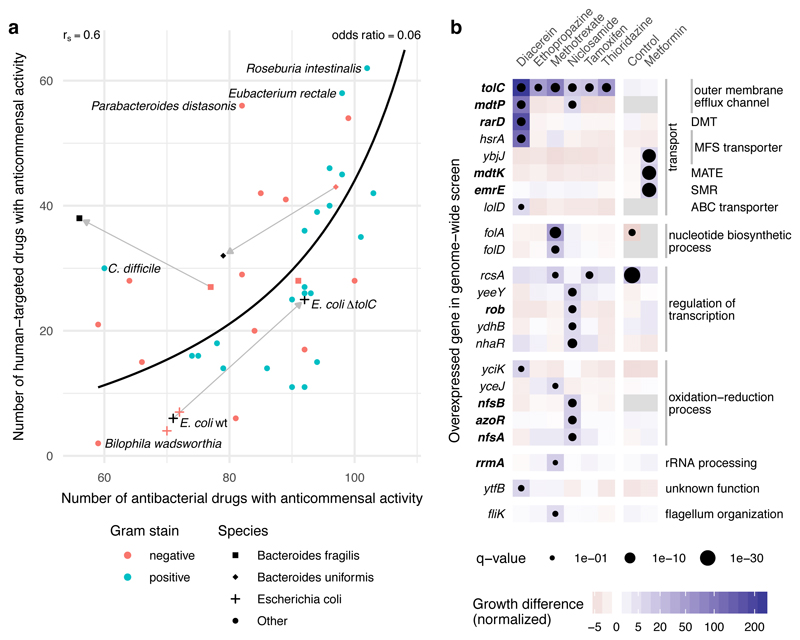
Figure 4. Antibiotic resistance mechanisms protect against human-targeted drugs.
a. Susceptibility to antibacterials and human-targeted drugs correlates across the 40 tested strains (Spearman correlation, rS=0.6 and a line depicting the nonlinear least-squares estimate of the odds ratio, OR=0.06), suggesting common resistance mechanisms against both drug types. Knocking out a major antibiotic efflux pump, tolC, in the lab E. coli strain, BW25113 (behaving as the other 2 commensal E. coli strains in the screen), makes E. coli equally more sensitive to both antibacterials and human-targeted drugs. Two antibiotic-resistant isolates of B. fragilis (black square, HM-20) and B. uniformis (black diamond, HM-715) were screened in addition to the main screen with only the latter showing a similar increase in resistance towards human-targeted drugs. b. Chemical genetic screen of an E. coli genome-wide overexpression library in 7 non-antibiotics; all screens except for metformin were performed in ΔtolC background to sensitize E. coli to these drugs. Genes that when overexpressed improved significantly the growth of E. coli to at least one of the drugs are shown here; in bold genes previously associated with antibiotic resistance. Among them, genes encoding for transporters from different families: DMT (drug metabolite transporter), MFS (major facilitator superfamily), MATE (multidrug and toxin extrusion), SMR (small multidrug resistance) and ABC (ATP-binding cassette). Growth is measured by colony size (median n=4) 41, color depicts the normalized size difference from the median growth of all strains in the drug (>6-fold difference), and dot size the significance (FDR-corrected p-value <0.1). Control denotes the growth of the library without drug.