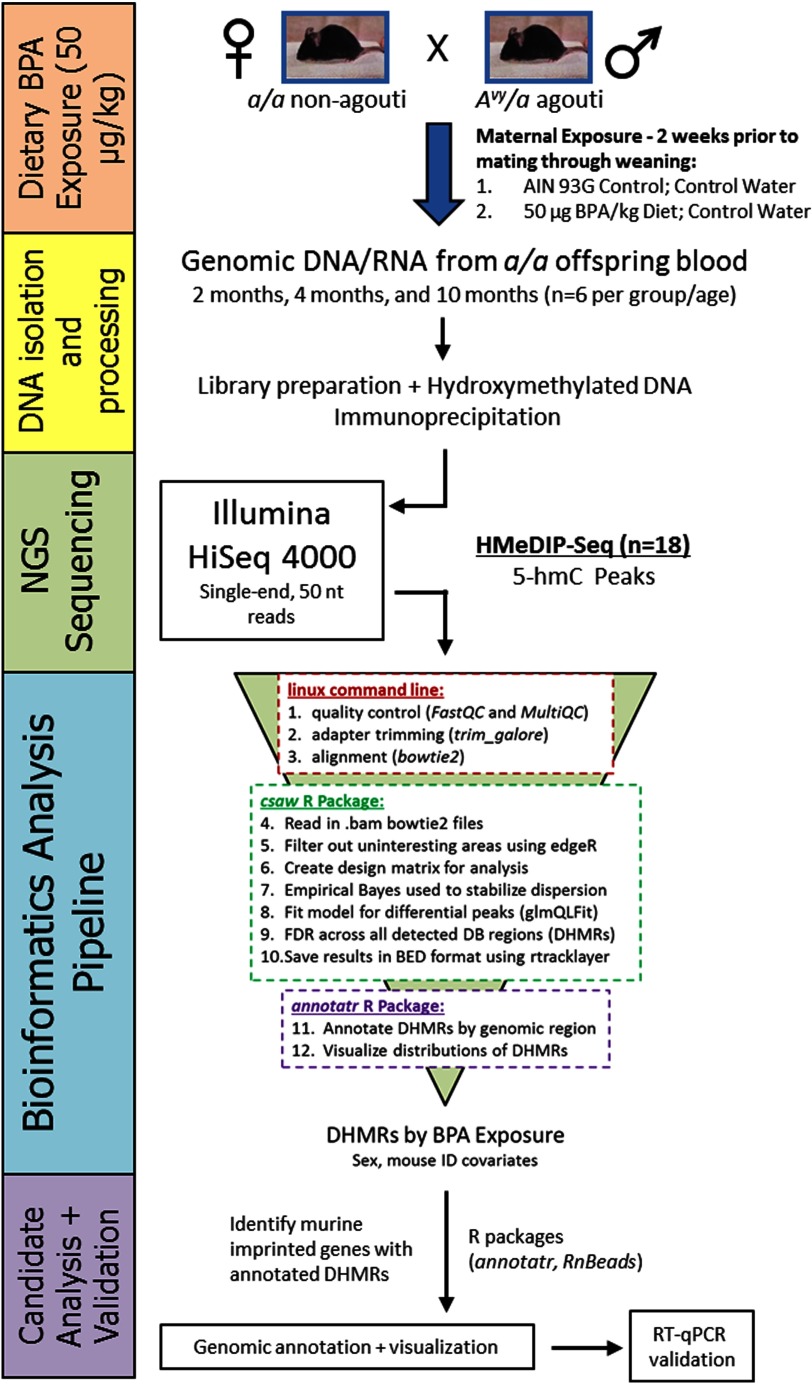
Figure 1.
Sequencing data collection and analysis workflow. Two weeks prior to mate-pairing with males, 8- to 10-wk-old wild-type dams were placed on one of two experimental diet groups: (1) Control (modified, phytoestrogen-free 7% corn oil AIN-93G), or (2) Control diet. Dietary exposure was continued through gestation and lactation until weaning at postnatal day 21. Genomic DNA was isolated from matched wild-type offspring blood samples at 2, 4, and 10 months of age. DNA was isolated from a subset of Control ( per age group) and BPA-exposed ( per age group) mice, then processed in preparation for HsMeDIP-seq. All processed samples were amplified and sequenced on an Illumina HiSeq 4000 sequencer using single-end, reads. BPA-related differentially hydroxymethylated regions (DHMRs) were identified and annotated using a bioinformatics pipeline. Annotated DHMRs were then visualized in the genome browser. The target gene region—Gnas—was then validated using RT-qPCR on available RNA from the blood samples.