Abstract
DNA crosslinking agents make up a broad class of chemotherapy agents that target rapidly dividing cancer cells by disrupting DNA synthesis. These drugs differ widely in both chemical structure and biological effect. In cells, crosslinking agents can form multiple types of DNA lesions with varying efficiencies. Inter-strand crosslinks (ICLs) are considered to be the most cytotoxic lesion, creating a covalent roadblock to replication and transcription. Despite over 50 years in the clinic, the use of crosslinking agents that specialize in the formation of ICLs remains limited, largely due to high toxicity in patients. Current ICL-based therapeutics have focused on late-stage and drug-resistant tumors, or localized treatments that limit exposure. In this article, we review the development of clinical crosslinking agents, our understanding of how cells respond to different lesions, and the potential to improve ICL-based chemotherapeutics in the future.
Keywords: chemotherapy, inter-strand crosslink (ICL), drug-resistance, DNA repair
Introduction
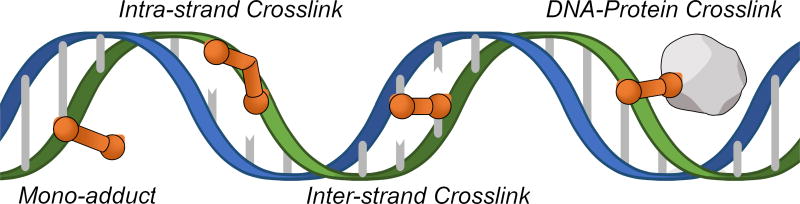
Crosslinking agents are a chemically diverse group of molecules that contain two or more reactive ends. In cells, the bi-functional nature of crosslinking agents can lead to covalent coupling between functional groups on DNA and other molecules. Reactions with DNA may involve one or both strands to form different types of lesions, including DNA mono-adducts, DNA-protein crosslinks, intra-strand crosslinks, and inter-strand crosslinks (ICLs) (Figure 1)[1]. Although crosslinking agents can produce a variety of DNA lesions in cells, cytotoxicity is often attributed to the formation of ICLs. By covalently coupling complementary strands of the DNA duplex, ICLs block strand separation that is required for fundamental cellular processes like DNA replication and transcription[2]. Failure to remove ICLs from DNA can block cell cycle progression and lead to cell death[3]. Defects in ICL repair can also lead to catastrophic chromosomal aberrations that promote tumorigenesis[4].
Figure 1. DNA lesions formed by crosslinking agents.
Crosslinking agents are highly reactive molecules that can form multiple types of DNA lesions in cells.
Congenital defects in ICL repair are associated with two major cancer predisposition syndromes, Fanconi anemia (FA) and hereditary breast and ovarian cancer susceptibility (BRCA). Together, FA and BRCA proteins form an adaptive DNA damage signaling and repair pathway that promotes removal of ICLs and restores the damaged DNA[5, 6]. The FA and BRCA pathways are highly inter-connected and share extensive genetic overlap[7, 8]. While FA-deficient cells are particularly sensitive to ICLs[5], BRCA mutants are sensitive to a range of DNA lesions, including ICLs[9].
The high cellular toxicity of ICLs has been exploited clinically by numerous anti-cancer therapies. Cancer is typified by rapid, uncontrolled cell proliferation. Like many chemotherapeutics, crosslinking agents selectively target cancer by interfering with DNA synthesis[10]. Although some crosslinking agents are used extensively in the clinic, those that form ICLs with high efficiency are typically reserved for late-stage and drug-resistant tumors[11], or localized treatments that reduce potential side effects[12–15]. In this review, we provide an update on several studies that shed new light on how cells respond to different ICL-inducing agents and their potential for future therapeutic application.
Diversity of Clinical Crosslinking Agents
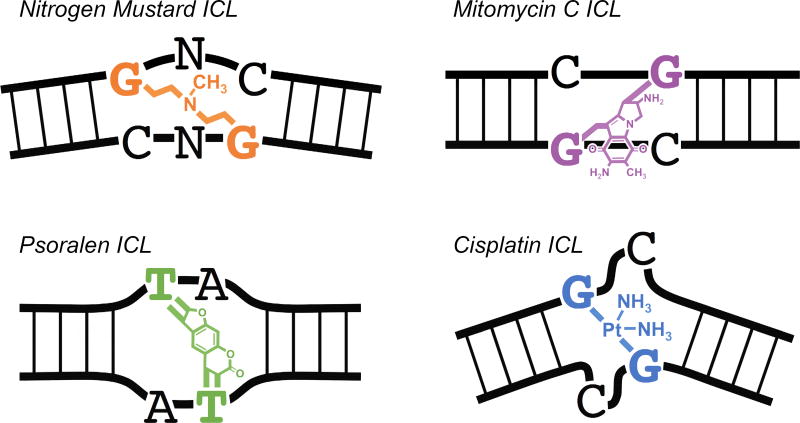
There are four major subgroups of crosslinking agents that have been developed for clinical use: nitrogen mustards, mitomycins, psoralens, and platinum-based compounds (Figure 2). Differences in chemical structure affect how these drugs interact with DNA and the type of lesions they create. The key features of each group are described below.
Figure 2. Diversity of ICL lesions.
There are four major subgroups of DNA crosslinking agents. The ICLs produced by each group can vary widely in chemical structure and their effect on DNA topology. Nitrogen Mustard ICLs cause mild DNA bending and unwinding. Mitomycin C ICLs cause minimal distortion to DNA. Psoralen ICLs cause moderate DNA unwinding. Cisplatin ICLs cause severe DNA bending and unwinding.
Nitrogen mustards
Nitrogen mustards are among the oldest and most extensively studied crosslinking agents[6, 16]. Several derivatives of nitrogen mustard (mechlorethamine) are still used regularly in the clinic, including melphalan and cyclophosphamide. Nitrogen mustards contain an N,N-bis-(2-chloroethyl) amine as the defining component. In cells, nitrogen mustards form mostly DNA mono-adducts and intra-strand crosslinks, with ICLs making up 5% or less of total lesions[16]. Nitrogen mustard ICLs are generated at the N7 of guanine in GpC (5’-G-phosphate-C-3’) or GpNpC DNA sequences[16]. Nitrogen mustard ICLs cause only limited DNA distortions, bending the duplex ~10 degrees and unwinding the helix by ~6 degrees[6].
Mitomycins
Mitomycins are a family of natural products that were originally isolated from Streptomyces caespitosis in the 1950s. Although several derivatives have been developed, Mitomycin C (MMC) remains the most clinically active. In cells, MMC can form up to 15% ICLs, in addition to a variety of DNA mono-adducts that make up over half of total lesions[6, 17]. Mitomycin C itself is only mildly reactive. In cells, MMC is “activated” by the reduction of its quinone moiety[11]. MMC ICLs are preferentially formed at CpG sequences[6, 11] and cause minimal distortion to the DNA helix[6, 18, 19].
Psoralens
Originally derived from plants, psoralen and derivatives like methoxypsoralen and trimethylpsoralen are photosensitive and become reactive when exposed to ultraviolet (UV) light[20]. Psoralens are planar in structure and able to intercalate between adjacent base pairs of duplex DNA. As a result, UV-activated psoralens readily form ICLs with high efficiency, ranging from ~40% with methoxypsoralen[21] to ~90% with trimethylpsoralen[6]. Psoralens preferentially react with TpA sequences in DNA to form covalent mono-adducts and ICLs. Psoralen ICLs do not bend the DNA duplex but do cause helical unwinding of ~25 degrees[6, 22, 23].
Platinums
A wide variety of platinum-based compounds have been developed for clinical use, including cisplatin, carboplatin, oxaliplatin, nedaplatin, and satraplatin. One of the most widely studied is cisplatin, which forms approximately 90% intra-strand crosslinks[24, 25] and only 1–2% ICLs in genomic DNA[26]. Cisplatin ICLs form almost exclusively at GpC sequences, reacting with the N7 of guanine[6]. Because cisplatin ICLs rest in the minor groove of DNA and force cytosine extrusion, these lesions cause severe distortion of DNA, bending the duplex ~47 degrees and unwinding the helix ~110 degrees[6].
Recognition and Repair of ICLs
The structure and position of an ICL play a major role in how the lesion is recognized and repaired in cells[27]. Various factors from different DNA repair pathways cooperate to recognize and remove ICLs from DNA, including those from nucleotide excision repair (NER), base excision repair (BER), mismatch repair (MMR), homologous recombination repair (HR), translesion synthesis (TLS), transcription coupled repair (TCR), and the FA/BRCA cancer predisposition pathways[6, 28]. Together, these pathways form an adaptive, non-linear stress response that coordinates removal of ICLs from DNA.
Replication-coupled repair
ICL repair occurs primarily during S phase after a replication fork collides with the crosslink[29, 30]. When replication forks are unable to bypass an ICL, a series of damage signaling events coordinate removal of the ICL, repair of the damaged DNA, and completion of DNA synthesis. For cisplatin ICLs, repair requires the convergence of two replication forks on the lesion[29]. In cells, the firing of dormant origins likely facilitates fork convergence when arrival of a neighboring fork is delayed[31]. Damage signaling pathways then promote dismantling of the replication machinery by the ubiquitin-selective p97 segregase, allowing repair enzymes to access the obstructed crosslink[32, 33]. Next, members of the FA/BRCA pathway, including FANCI-FANCD2[34], promote DNA incision by XPF-ERCC1 that “unhook” the ICL from one DNA strand[35, 36]. The resulting DNA double-strand break is repaired by homologous recombination[37] and the remaining ICL adduct is likely removed by NER.
ICL traverse
Replication forks have also been shown to “traverse” a trimethylpsoralen ICL, continuing DNA synthesis past the crosslink without repair[38]. This process is supported by FANCM, a DNA translocase that interacts with the replisome through the processivity factor PCNA[39]. Although the mechanism remains unclear, ICL traverse allows cells to complete DNA synthesis and repair the ICL at a later time, likely using a replication-independent mechanism described below.
Direct cleavage of psoralen ICLs
Unlike ICLs generated by other agents, psoralen crosslinks contain an N-glycosyl bond that can be cleaved directly by NER DNA glycosylases like NEIL1[40, 41] and NEIL3[42, 43]. Cleavage of the glycosidic bond does not require displacement of the replicative helicase and is not dependent on FANCI-FANCD2 or the associated nucleases[34, 42]. Thus, psoralen ICLs can be readily bypassed without the formation of a highly toxic DNA double-strand break. As with replication-coupled repair, the remaining ICL adducts are likely removed by NER.
Replication-independent repair
Outside of S phase, there are at least two distinct mechanisms for ICL recognition. First, global genome surveillance proteins like XPC-HR23B and DDB1-DDB2 are thought to recognize helical distortions caused by some ICLs[6, 44, 45]. Second, transcription of cross-linked DNA can lead to stalling of RNA polymerase[46, 47]. In both cases, subsequent repair is dependent on NER and TLS proteins[44, 46, 47]. After incisions unhook the ICL from one DNA strand, TLS polymerases fill in the single-stranded gap. The unhooked adduct is then removed from the other DNA strand by a second round of incisions and gap filling.
Targeting cancer with ICL therapeutics
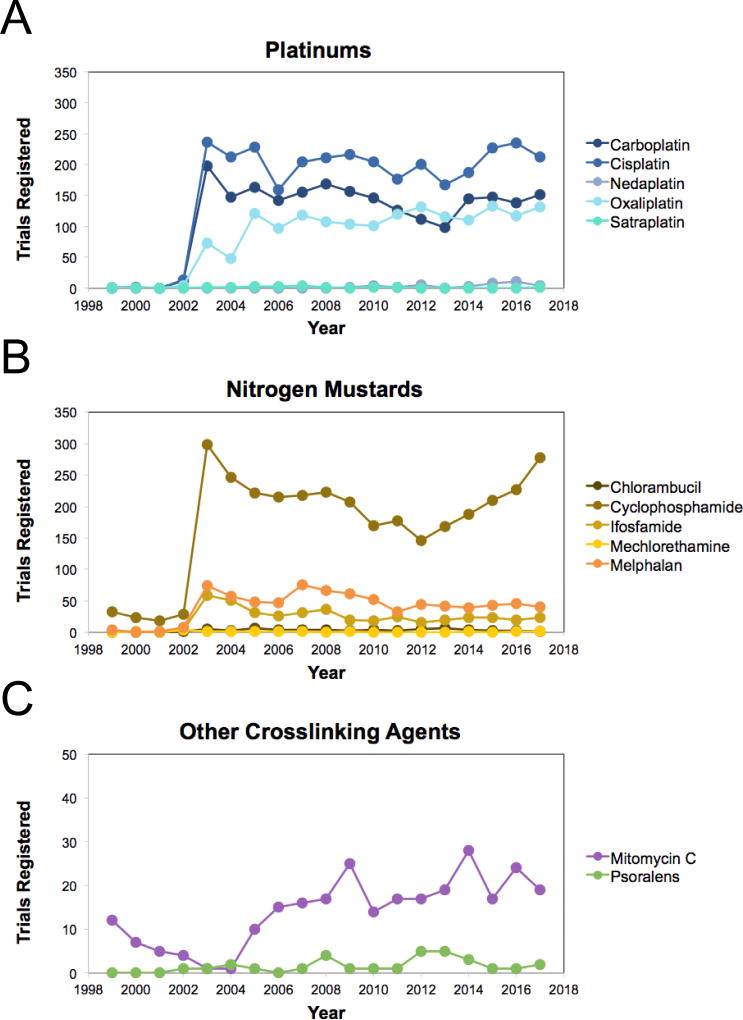
Because of their potent anti-cancer properties, DNA crosslinking agents have remained at the frontline of chemotherapy for more than 50 years. There are currently over 4,400 open clinical trials that utilize crosslinking agents (Table 1). About a quarter of these are late-phase trials (phase III or IV) that have proven to be effective in early studies and show the most potential to improve the standard of care. Over half of all open crosslink trials utilize either cisplatin (~23%) or the nitrogen mustard derivative cyclophosphamide (~28%). However, clinical use of crosslinking agents that create ICLs with high efficiency remains limited due to high toxicity associated with treatment[48]. Together, MMC and psoralen compounds make up only 2% of open clinical trials, with the number of new registrations relatively unchanged over the last 12 years (Figure 3C). Thus, most crosslink-based treatments are not currently designed to create ICLs, but instead heavily favor the formation of other types of DNA lesions.
Table 1.
Open clinical trials with DNA crosslinking agents. The number of open clinical trials registered with clinicaltrials.gov is listed for crosslinking agents from each major subgroup. ‘Total’ includes trials that are active and currently recruiting, enrolling by invitation, or active but not recruiting. ‘Late Phase’ is defined as phase III and phase IV trials. ‘Combo’ represents the number of late-phase trials that combine a DNA crosslinking agent with at least one additional chemotherapeutic agent. ‘Resistant’ represents the number of late-phase trials that target drug-resistant, refractory, relapsed, or recurrent cancers unresponsive to previous therapy. All data current through 01/ 01/2018.
| Late phase
|
|||||
|---|---|---|---|---|---|
| Name | Total | Total | Combo | Resistant | Conditions treated |
| Platinums | |||||
| Cisplatin | 1035 | 315 | 299 | 29 | Non-small cell lung cancer, hepatocellular carcinoma, biliary tract cancer, mesothelioma, breast cancer, brain neoplasms, and gynecological cancers |
| Carboplatin | 748 | 197 | 197 | 44 | Abdominal and rectal cancers, liver cancer, esophageal cancer, neuroblastoma, pancreatic cancer, biliary tract cancer, hepatocellular carcinoma, lung cancer, breast cancer, and gynecological cancers |
| Oxaliplatin | 574 | 168 | 168 | 11 | Digestive system cancer, liver cancer, esophageal cancer, neuroblastoma, pancreatic cancer, peritoneal carcinomatosis, bile duct cancer, biliary tract cancer, hepatocellular carcinoma, and lung cancer |
| Nedaplatin | 18 | 6 | 6 | – | Nasopharyngeal, esophageal cancer, and non-small cell lung cancer |
| Satraplatin | 1 | – | – | – | Prostate cancer |
| Nitrogen mustards | |||||
| Cyclophosphamide | 1222 | 264 | 257 | 22 | Crohn’s disease, neuroblastoma, sarcomas, breast cancer, endometrial cancer, leukemia, lymphoma, myeloma, thyroid cancer, systemic sclerosis, and lupus nephritis |
| Ifosfamide | 428 | 30 | 28 | 4 | Small cell lung cancer, soft tissue sarcoma, lymphoma, osteosarcoma, germ cell tumor, nasopharynx carcinoma, bladder cancer, testicular cancer, squamous cell carcinoma of the penis, cervical cancer, pleuropulmonary blastoma, neuroblastoma, unresectable sinonasal tumors, and pancreatic cancer |
| Melphalan | 268 | 40 | 40 | 7 | Metastatic melanoma, multiple myeloma, hematopoietic stem cell transplantation, prostate cancer, colorectal cancer, lymphomas, brain neoplasms, leukemia, retinoblastoma, bile duct cancer, spinal tumors, autoimmune disease, sickle cell disease, testicular cancer, myelofibrosis, Ewing’s sarcoma, and amyloidosis |
| Chlorambucil | 18 | 9 | 7 | 1 | Chronic lymphocytic leukemia, extranodal MALT lymphoma, small cell lymphoma, and bladder cancer |
| Mechlorethamine | 9 | 2 | 2 | – | Mycosis fungoides, cutaneous T-cell lymphoma, and Hodgkin’s lymphoma |
| Other | |||||
| Mitomycin C | 89 | 30 | 13 | 5 | Glaucoma, bladder cancer, colorectal cancer, anal cancer, pancreatic cancer, bile duct cancer, gastric cancer, liver cancer, head and neck cancers, peritoneal cancer, gynecological cancers, mesothelioma, gastrointestinal cancer, metastatic breast cancer, pterygium, presbyopia, corneal opacity, obstructive airway disease, and post-LASIK keratectasia |
| Psoralens | 4 | 1 | – | – | Vitiligo and cutaneous T-cell lymphoma |
Figure 3. Clinical use of DNA crosslinking agents over time.
The number of clinical trials registered each year with ClinicalTrials.gov is graphed for the most commonly used crosslinking agents from each major subgroup. ClinicalTrials.gov consists of privately and publicly funded clinical studies conducted around the world. The database was established by the Food and Drug Administration Modernization Act of 1997 (FDAMA) and is maintained by the National Library of Medicine (NLM) at the National Institutes of Health (NIH).
In the clinic, crosslinking agents are frequently used to induce cellular stress and enhance the cytotoxicity of other agents. In late-phase trials, over 95% of crosslink-based therapies involve combinations with other chemotherapeutic agents (Table 1). Combinational treatments typically administer drugs at lower doses than single-drug therapies, which can reduce toxic side effects associated with treatment. Combining drugs that act through different mechanisms is also an established strategy for preventing the development of drug resistance, which remains one of the primary challenges for anti-cancer chemotherapies. Although many tumors are initially sensitive to treatment, cancer genomes are inherently unstable and can develop tolerance through several mechanisms. Cells can acquire both genetic and epigenetic changes that disrupt drug-target interactions, increase drug efflux from cells, and alter cellular signaling pathways that control the DNA damage response and cell death[49, 50]. On average, less than 12% of late-phase trials that utilize crosslinking agents target unresponsive, relapsed, or recurrent cancers, emphasizing the need to develop additional therapies that can treat drug-resistant tumors.
To combat both innate tolerance and acquired resistance to chemotherapeutics, many treatment strategies seek to exploit changes in cancer physiology. For example, cancer cells frequently acquire defects in DNA repair during both the development and progression of cancer[53]. Consequently, many tumors are highly sensitive to different types of DNA damage. In order to predict which tumors will be sensitive to treatment, a variety of biomarkers from different DNA repair pathways have been identified, including BRCA1, BRCA2, ATM, ATR, CHK1/2, and FANCD2[54–56]. Using patient-derived tumor samples, changes in gene sequence, protein expression, and post-translational modification have been shown to strongly correlate with cellular sensitivity to specific DNA damaging agents[57]. However, the impact that many biomarkers have in the clinic is limited by high analytical costs and a lack of viable treatment options. As a result, many treatments are administered without stratifying patients based on DNA repair capabilities.
Going forward, additional therapeutic strategies are needed to harness the potent cytotoxic effects of ICLs. Recent studies have revealed a diverse range of repair mechanisms utilized by cells in response to different ICL lesions. New roles have been identified for proteins like NEIL1/3, p97, and FANCM during ICL repair, highlighting these factors and their respective pathways as valuable targets for combinational therapy with crosslinking agents. With the advent of tumor profiling, novel molecular inhibitors, and a growing list of DNA damage response biomarkers, there is immense potential to exploit these mechanistic observations clinically. Establishing highly selective and targeted therapies will not only improve the efficacy of current ICL-based treatments, but also allow them to be applied more broadly by reducing the toxic side effects that serve as a barrier to widespread use.
Highlights.
DNA crosslinking agents have been widely used in the clinic for over 50 years.
Crosslinking agents can generate different DNA lesions with varying efficiencies.
Due to high toxicity, most crosslink-based therapies avoid formation of ICLs.
New mechanistic observations highlight promise for exploiting the potent effects of ICLs.
Acknowledgments
Funding
This work was supported by the National Institutes of Health (R35GM119512 to D.T.L.) and the NIH National Center for Advancing Translational Sciences (TL1 TR001451 and UL1 TR001450 to H.B.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brulikova L, Hlavac J, Hradil P. DNA interstrand cross-linking agents and their chemotherapeutic potential. Curr Med Chem. 2012;19(3):364–85. doi: 10.2174/092986712803414295. [DOI] [PubMed] [Google Scholar]
- 2.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493(7432):356–63. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osawa T, Davies D, Hartley JA. Mechanism of cell death resulting from DNA interstrand cross-linking in mammalian cells. Cell Death &Amp; Disease. 2011;2:e187. doi: 10.1038/cddis.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12(12):801–17. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 5.Duxin JP, Walter JC. What is the DNA repair defect underlying Fanconi anemia? Curr Opin Cell Biol. 2015;37:49–60. doi: 10.1016/j.ceb.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez-Martinez D, Liang CC, Cohn MA. Cellular response to DNA interstrand crosslinks: the Fanconi anemia pathway. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2218-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sawyer SL, et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5(2):135–42. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Andrea AD. BRCA1: A Missing Link in the Fanconi Anemia/BRCA Pathway. Cancer discovery. 2013;3(4):376–378. doi: 10.1158/2159-8290.CD-13-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 10.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nature reviews. Cancer. 2011;11(7):467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomasz M. Mitomycin C: small, fast and deadly (but very selective) Chem Biol. 1995;2(9):575–9. doi: 10.1016/1074-5521(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 12.Serretta V, et al. Mitomycin C from birth to adulthood. Urologia. 2016;83(Suppl 2):2–6. doi: 10.5301/uro.5000195. [DOI] [PubMed] [Google Scholar]
- 13.Ragonese M, et al. Mitomycin C: new strategies to improve efficacy of a well-known therapy. Urologia. 2016;83(Suppl 2):24–28. doi: 10.5301/uro.5000193. [DOI] [PubMed] [Google Scholar]
- 14.Whitton ME, et al. Interventions for vitiligo. Cochrane Database Syst Rev. 2015;(2):Cd003263. doi: 10.1002/14651858.CD003263.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan WH, et al. Juvenile mycosis fungoides with large-cell transformation: Successful treatment with psoralen with ultraviolet A light, interferon-alfa, and localized radiation. Pediatr Dermatol. 2017 doi: 10.1111/pde.13338. [DOI] [PubMed] [Google Scholar]
- 16.Rajski SR, Williams RM. DNA Cross-Linking Agents as Antitumor Drugs. Chem Rev. 1998;98(8):2723–2796. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- 17.Warren AJ, Maccubbin AE, Hamilton JW. Detection of Mitomycin C-DNA Adducts <em>in Vivo</em> by <sup>32</sup>P-Postlabeling: Time Course for Formation and Removal of Adducts and Biochemical Modulation. Cancer Research. 1998;58(3):453–461. [PubMed] [Google Scholar]
- 18.Rink SM, et al. Bending of DNA by the mitomycin C-induced, GpG intrastrand cross-link. Chem Res Toxicol. 1996;9 doi: 10.1021/tx950156q. [DOI] [PubMed] [Google Scholar]
- 19.Tomasz M, et al. Isolation and structure of a covalent cross-link adduct between mitomycin C and DNA. Science. 1987;235(4793):1204. doi: 10.1126/science.3103215. [DOI] [PubMed] [Google Scholar]
- 20.Isaacs ST, et al. Synthesis and characterization of new psoralen derivatives with superior photoreactivity with DNA and RNA. Biochemistry. 1977;16(6):1058–1064. doi: 10.1021/bi00625a005. [DOI] [PubMed] [Google Scholar]
- 21.Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat Res. 2001;486 doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 22.Spielmann HP, et al. Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics. Biochemistry. 1995;34(40):12937–53. [PubMed] [Google Scholar]
- 23.Haran TE, Crothers DM. Phased psoralen cross-links do not bend the DNA double helix. Biochemistry. 1988;27 doi: 10.1021/bi00418a044. [DOI] [PubMed] [Google Scholar]
- 24.Eastman A. The formation, isolation and characterization of DNA adducts produced by anticancer platinum complexes. Pharmacol Ther. 1987;34(2):155–66. doi: 10.1016/0163-7258(87)90009-x. [DOI] [PubMed] [Google Scholar]
- 25.Kasparkova J, Brabec V. Recognition of DNA interstrand cross-links of cisdiamminedichloroplatinum( II) and its trans isomer by DNA-binding proteins. Biochemistry. 1995;34(38):12379–87. doi: 10.1021/bi00038a035. [DOI] [PubMed] [Google Scholar]
- 26.Pera MF, et al. Quantitative aspects of the formation and loss of DNA interstrand crosslinks in Chinese hamster cells following treatment with cis-diamminedichloroplatinum(II) (cisplatin) II. Comparison of results from alkaline elution, DNA renaturation and DNA sedimentation studies. Biochimica et Biophysica Acta (BBA) - Nucleic Acids and Protein Synthesis. 1981;655(2):152–166. doi: 10.1016/0005-2787(81)90005-8. [DOI] [PubMed] [Google Scholar]
- 27.Beljanski V, Marzilli LG, Doetsch PW. DNA Damage-Processing Pathways Involved in the Eukaryotic Cellular Response to Anticancer DNA Cross-Linking Drugs. Molecular Pharmacology. 2004;65(6):1496–1506. doi: 10.1124/mol.65.6.1496. [DOI] [PubMed] [Google Scholar]
- 28.Nepal M, et al. Fanconi Anemia Signaling and Cancer. Trends Cancer. 2017;3(12):840–856. doi: 10.1016/j.trecan.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, et al. DNA interstrand cross-link repair requires replication-fork convergence. Nat Struct Mol Biol. 2015;22(3):242–7. doi: 10.1038/nsmb.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akkari YM, et al. DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol Cell Biol. 2000;20(21):8283–9. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y-H, et al. ATR-Mediated Phosphorylation of FANCI Regulates Dormant Origin Firing in Response to Replication Stress. Molecular Cell. 2015;58(2):323–338. doi: 10.1016/j.molcel.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •32.Fullbright G, et al. p97 Promotes a Conserved Mechanism of Helicase Unloading during DNA Cross-Link Repair. Mol Cell Biol. 2016;36(23):2983–2994. doi: 10.1128/MCB.00434-16. This study describes how the p97 segregase recognizes and removes the CMG replicative helicase from DNA after it collides with a DNA inter-strand crosslink. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long DT, et al. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol Cell. 2014;56(1):174–85. doi: 10.1016/j.molcel.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knipscheer P, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326 doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •35.Abdullah UB, et al. RPA activates the XPF•ERCC1 endonuclease to initiate ERCC1 endonuclease to initiate processiing of DNA interstrand crosslinks. The EMBO Journal. 2017;36(14):2047–2060. doi: 10.15252/embj.201796664. In this study, the authors show that binding of RPA to single-stranded DNA stimulates processing of ICLs by the XPf-ERCC1 endonuclease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein Douwel D, et al. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 2014;54(3):460–71. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long DT, et al. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333 doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang J, et al. The DNA Translocase FANCM/MHF Promotes Replication Traverse of DNA Interstrand Crosslinks. Molecular cell. 2013;52(3) doi: 10.1016/j.molcel.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •39.Rohleder F, et al. FANCM interacts with PCNA to promote replication traverse of DNA interstrand crosslinks. Nucleic Acids Research. 2016;44(7):3219–3232. doi: 10.1093/nar/gkw037. The authors provide a link between FANCM and the replisome by showing it interacts with PCNA through a PIP-box binding domain in response to replication stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Couvé S, et al. The Human Oxidative DNA Glycosylase NEIL1 Excises Psoralen-induced Interstrand DNA Cross-links in a Three-stranded DNA Structure. The Journal of Biological Chemistry. 2009;284(18):11963–11970. doi: 10.1074/jbc.M900746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McNeill DR, et al. NEIL1 responds and binds to psoralen-induced DNA interstrand crosslinks. J Biol Chem. 2013;288(18):12426–36. doi: 10.1074/jbc.M113.456087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •42.Semlow DR, et al. Replication-Dependent Unhooking of DNA Interstrand Cross-Links by the NEIL3 Glycosylase. Cell. 2016;167(2):498–511. e14. doi: 10.1016/j.cell.2016.09.008. The authors show that the NEIL3 glycosylase cleaves a psoralen ICL, promoting replication bypass in the absence of FANCI-FANCD2-mediated DNA incision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z, et al. A role for the base excision repair enzyme NEIL3 in replication-dependent repair of interstrand DNA cross-links derived from psoralen and abasic sites. DNA Repair (Amst) 2017;52:1–11. doi: 10.1016/j.dnarep.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isaacs RJ, Spielmann HP. A model for initial DNA lesion recognition by NER and MMR based on local conformational flexibility. DNA Repair (Amst) 2004;3(5):455–64. doi: 10.1016/j.dnarep.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 45.Muniandy P, et al. DNA INTERSTRAND CROSSLINK REPAIR IN MAMMALIAN CELLS: STEP BY STEP. Crit Rev Biochem Mol Biol. 2010;45(1):23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enoiu M, Jiricny J, Scharer OD. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res. 2012;40(18):8953–64. doi: 10.1093/nar/gks670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muniandy PA, et al. Repair of laser-localized DNA interstrand cross-links in G1 phase mammalian cells. J Biol Chem. 2009;284(41):27908–17. doi: 10.1074/jbc.M109.029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verweij J, Pinedo HM. Mitomycin C: mechanism of action, usefulness and limitations. Anticancer Drugs. 1990;1(1):5–13. [PubMed] [Google Scholar]
- 49.Kibria G, Hatakeyama H, Harashima H. Cancer multidrug resistance: mechanisms involved and strategies for circumvention using a drug delivery system. Arch Pharm Res. 2014;37(1):4–15. doi: 10.1007/s12272-013-0276-2. [DOI] [PubMed] [Google Scholar]
- 50.Alfarouk KO, et al. Resistance to cancer chemotherapy: failure in drug response from ADME to P-gp. Cancer Cell International. 2015;15:71. doi: 10.1186/s12935-015-0221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen D-W, et al. Cisplatin Resistance: A Cellular Self-Defense Mechanism Resulting from Multiple Epigenetic and Genetic Changes. Pharmacological Reviews. 2012;64(3):706–721. doi: 10.1124/pr.111.005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Norquist B, et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol. 2011;29(22):3008–15. doi: 10.1200/JCO.2010.34.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nickoloff JA, et al. Drugging the Cancers Addicted to DNA Repair. JNCI Journal of the National Cancer Institute. 2017;109(11):djx059. doi: 10.1093/jnci/djx059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •54.Stover EH, et al. Biomarkers of Response and Resistance to DNA Repair Targeted Therapies. Clin Cancer Res. 2016;22(23):5651–5660. doi: 10.1158/1078-0432.CCR-16-0247. A recent review describing current DNA repair biomarkers and their relationship to different anti-cancer chemotherapeutics. [DOI] [PubMed] [Google Scholar]
- 55.Velic D, et al. DNA Damage Signalling and Repair Inhibitors: The Long-Sought-After Achilles’ Heel of Cancer. Biomolecules. 2015;5(4):3204–3259. doi: 10.3390/biom5043204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Carrigan B, et al. Phase I trial of a first-in-class ATR inhibitor VX-970 as monotherapy (mono) or in combination (combo) with carboplatin (CP) incorporating pharmacodynamics (PD) studies. Journal of Clinical Oncology. 2016;34(15_suppl):2504–2504. [Google Scholar]
- •57.Martens-de Kemp SR, et al. The FA/BRCA Pathway Identified as the Major Predictor of Cisplatin Response in Head and Neck Cancer by Functional Genomics. Mol Cancer Ther. 2017;16(3):540–550. doi: 10.1158/1535-7163.MCT-16-0457. The authors show that knock down of various FA/BRCA proteins by siRNA increases sensitivity to cisplatin in different HNSCC cell lines. [DOI] [PubMed] [Google Scholar]