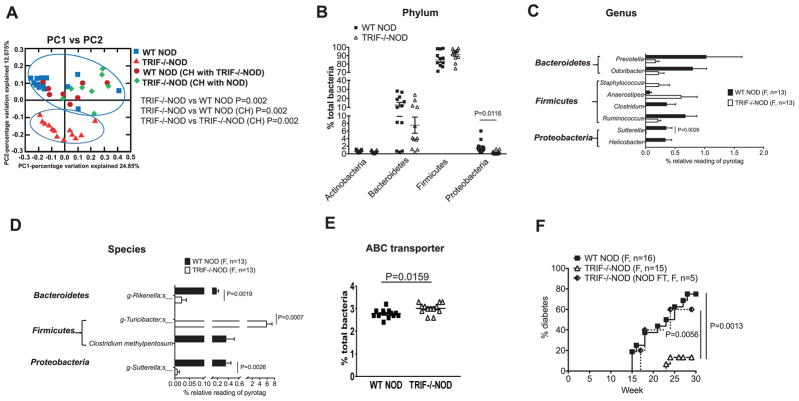
Fig. 2.
TRIF deficiency alters the gut microbiota in NOD mice. Fecal samples from female WT NOD and TRIF−/−NOD mice were used for taxonomic analysis by 16S rRNA sequencing. (A) PCoA plot showing β-diversity. ANOSIM was used for statistical analysis. Significant taxonomic compositions of the gut microbiota of non-cohoused WT NOD and TRIF−/−NOD mice were shown by as phylum (B), genus (C) and species (D). (E) Predictive functional profiling illustrating increased expression of ABC transporter in TRIF−/−NOD mice. (F) Diabetes incidence from TRIF−/−NOD mice that were gavaged with a suspension of NOD feces (NOD FT) compared with incidence in WT NOD and TRIF−/−NOD mice. The functional composition of the metagenome of the bacteria communities was predicted using PICRUSt. Multiple t tests with Bonferroni correction were used for statistical analysis in (B–D) while a two-tailed Student’s t test was used for statistical analysis in (E) and a log-rank (mantel-cox) test was used in (F).