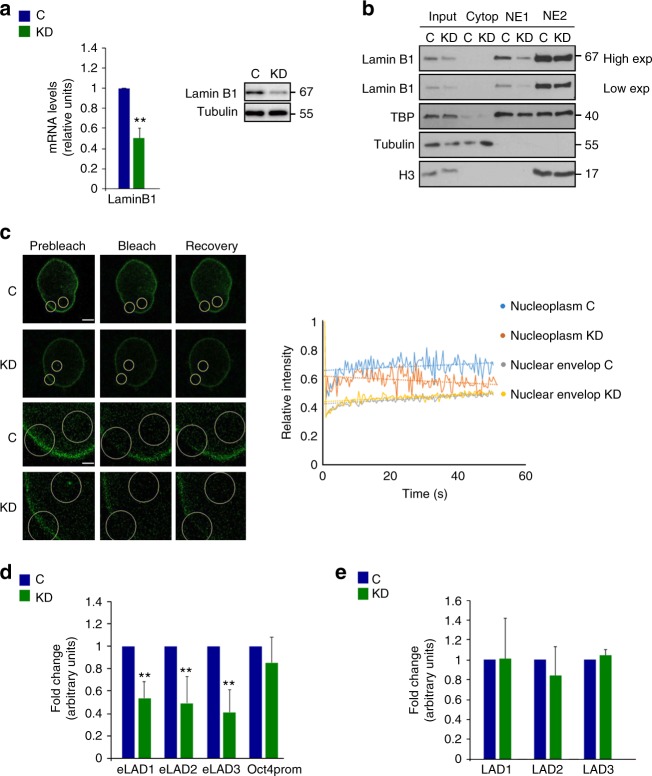
Fig. 5.
Knockdown of lamin B1 mainly affects non-NL integrated lamin B1. a qRT-PCR showing mRNA levels of lamin B1 in KD NMuMG cells. Expression levels were normalized to an endogenous control and expressed relative to the control-infected cells (C), which was set as 1. Data are presented as mean ± SD, n = 5. Left panel, representative western blotting for LB1 and tubulin (as a loading control) from C and KD NMuMG cells. Images are representative of three independent replicates (right). b Fractionation of nuclei from C and KD NMuMG cells to obtain cytoplasmic proteins, soluble proteins (NE1), and insoluble proteins (NE2), as monitored by western blotting using TBP as a NE1 control, tubulin as a cytoplasmic control, and H3 as a NE2 control. Images are representative of two independent replicates and five technical replicates. c Confocal fluorescence recovery after bleaching (FRAP) in C and KD cells expressing lamin B1-mCerulean. Circles denote the areas of bleaching. Each column displays prebleach, bleach, and recovery images. Rows 3 and 4 are magnifications of rows 1 and 2, respectively. Scale bar: 5 μm (inset 1 μm) (left). Images are representative of ten cells for each condition. Graphic representation of the relative intensity after 60 s of nuclear envelope and nucleoplasm recovery from bleaching in C and KD cells (right). Data are shown as mean, n = 6. d ChIP-qPCR of three selected lamin B1 + regions and a negative control (Oct4prom) selected from a negative lamin B1 + region, in C and KD NMuMG cells. Data from real-time PCR (qPCR) amplifications were normalized to the input and expressed as the fold-change relative to data obtained in C condition, which was set as 1. Data are shown as mean ± SD, n = 5. e ChIP-qPCR of three cLAD-selected regions in C and KD NMuMG cells. Data from qPCR amplifications were normalized to the input and expressed as the fold-change relative to data obtained in C condition, which was set as 1. Data presented as mean ± SD, n = 3