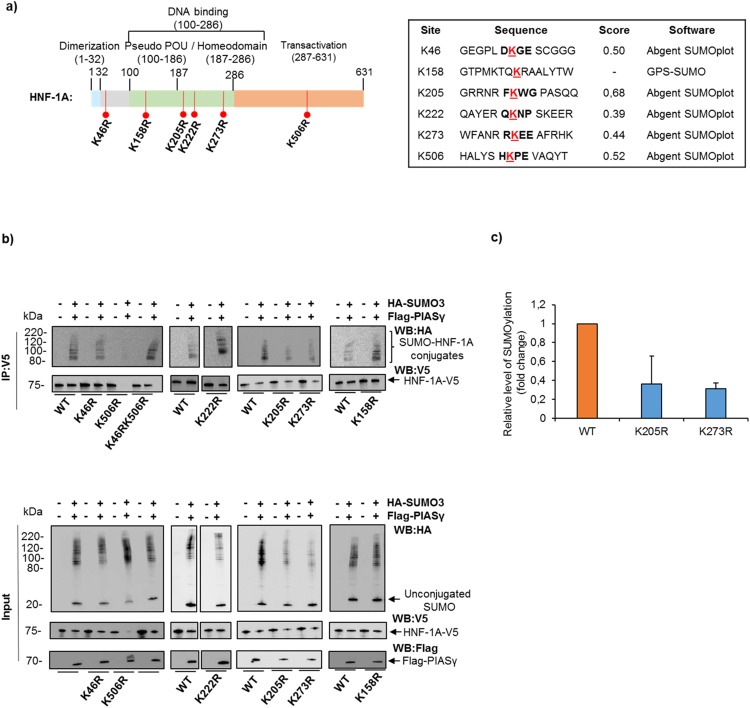
Figure 2.
K205 and K273 are SUMOylation sites in HNF-1A. (a) Schematic overview of the HNF-1A protein highlighting the lysine residues predicted as SUMOylation sites using the in silico prediction programs SUMOplot and GPS-SUMO. The predicted motif for covalent SUMO attachment is displayed with the lysine residue underlined in red. (b) Site directed mutagenesis of HNF-1A demonstrated loss of SUMOylation upon substitution lysine (K) with arginine (R), at residues K205, K273 and K506 in HEK293 cells. Cells were transfected with V5-tagged HNF-1A (WT or mutants) together with HA-tagged SUMO-3 and Flag-tagged PIASγ. Lysates were collected in the presence of NEM and protease inhibitors and subjected to immunoprecipitation using anti-V5 antibody. The precipitates were separated by SDS-PAGE and immunoblotting using anti-HA and anti-V5 antibodies. Full-length blots are presented in Supplementary Fig. S5. This experiment was replicated in three independent experimental days (n = 3). (c) Quantification of high molecular SUMOylated bands by densiometric analysis is presented as relative fold change compared to WT alone (set to 1). Each bar represents a mean of three independent experiments ± SD (n = 3).