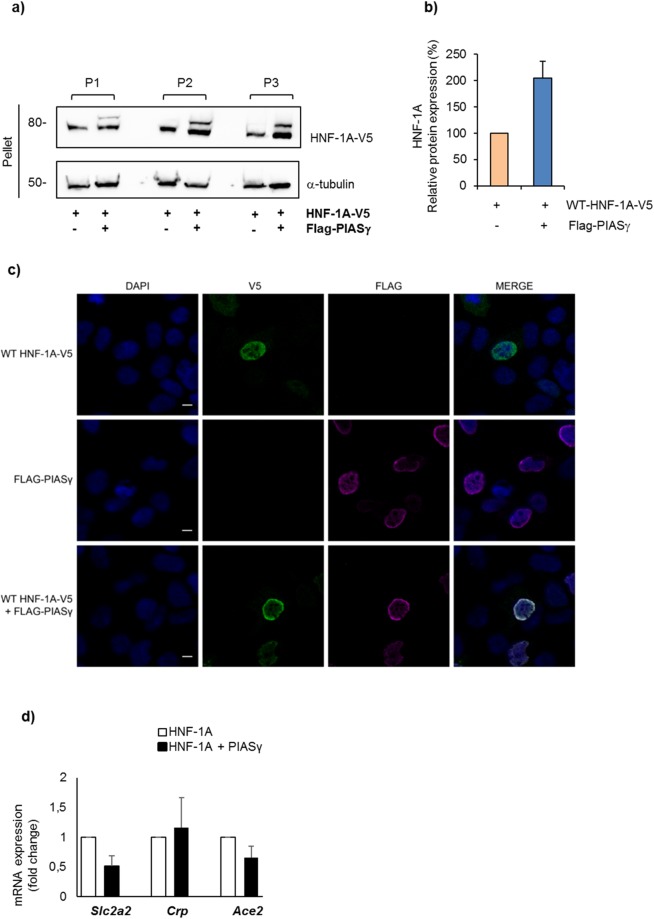
Figure 8.
PIASγ translocates HNF-1A to the nuclear periphery and leads to reduced expression of known HNF-1A target genes. (a,b) After RIPA lysis, pelleted fractions from HEK293 cells transiently transfected with V5-tagged HNF-1A in the absence/presence of PIASγ were analyzed by SDS-PAGE and immunoblotting using indicated antibodies. The level of HNF-1A was quantified by densiometric analysis using the Image Lab v.6.0.0 software (BIO-RAD) and normalized to α-tubulin. Full-length blots are presented in Supplementary Fig. S11. Quantifications are presented relative to the level of HNF-1A alone. The results are shown as mean of three technical experiments (n = 1) ±SD. P indicates parallel. (c) HEK293 cells were transiently transfected with constructs expressing V5-tagged HNF-1A and/or Flag-PIASγ alone or in combination as indicated in the figure. HNF-1A (green) and PIASγ (magenta) were visualized by immunostaining for V5- and Flag-tag, respectively, and analyzed by confocal microscopy. Cell nuclei were stained with DAPI (blue). Co-localization of transfected HNF-1A and PIASγ is shown in the lower panel. Scalebar, 10 µm. (d) The mRNA expression of three known HNF1A targets genes (Ace2, Slc2a2 and Crp) was investigated in MIN6 cells overexpressed with HNF1A in the absence or presence of PIASy and was determined by qPCR. Target gene expression was normalized to reference gene Gapdh and represented as mean fold change relative to HNF1A alone ±SD of two biological replicates (n = 2).