Abstract
A lipase producing strain B1213 isolated from soil was identified as Burkholderia pyrrocinia based on 16S rRNA gene and recA sequeence analysis, making this the first report on the presence of a lipase from B. pyrrocinia. Under an aqueous two-phase purification strategy, which included (ATPE)-ion-exchange chromatography (IEC)-gel and filtration chromatography (GFC), the specific activity of the 35-kDa lipase was determined to be 875.7 U/mg protein. The optimum pH and temperature of this lipase was pH 8.0 and 50 °C, respectively. The lipase retained > 85% activity in isopropanol and acetone at 30 °C for 10 min but the activity was reduced to 10.6% in n-hexane. Mg2+, Al3+, Mn2+, and Fe3+ enhanced lipase activity at both 1 mM and 5 mM concentrations. p-NPP, a long-chain acyl group 4-NP ester, appeared to be a good substrate candidate.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1414-9) contains supplementary material, which is available to authorized users.
Keywords: B. pyrrocinia, Lipase, Screening, Purification, Characterization
Introduction
Lipases are triacylglycerol ester hydrolases (EC 3.1.1.3) that have been widely used in biocatalysis due to their strong ability to catalyze the hydrolysis of triacylglycerides in aqueous solutions and in synthetic reactions (Feng et al. 2013; Gupta et al. 2004; Masomian et al. 2013), including hydrolysis, synthesis, inter-esterification, alcoholysis, acidolysis, esterification, aminolysis, acylation, and the resolution of racemates (Gupta et al. 2004; Gutarra et al. 2009; Salihu and Alam 2015; Yang et al. 2010). Lipases from various sources perform differently in terms of pH stability, organic solvent tolerance, and cold activity, and thus play very important roles in transforming various organic materials and biomass into useful products under conditions that are considered unsuitable to other biomolecules (de Abreu et al. 2014; Joseph et al. 2011; Maiangwa et al. 2015). Therefore, lipases have been widely used in many industrial applications, especially in organic synthetic compounds, detergents, food, feeds, paper industries, biodiesel production perfumes, cosmetics, leathers, enantiopure pharmaceuticals, and medical diagnostics (Aguieiras et al. 2015; Hasan et al. 2006; Masomian et al. 2013).
Lipases are ubiquitous in nature and can be found in most animals and plants, and most of them thrive in microorganisms (Gupta et al. 2004, 2015; Nagarajan 2012). The action of microbial lipases widely depends on temperature, pH, and substrate specificity, which can result in short reaction times, parameters that are important in industrial applications, such as food processing industries, synthesis of fine chemicals, biodiesel, etc. In the entire microbial lipase family, most enzymes in the biotechnological applications and organic chemistry fields are from natural bacterial and fungal, as well as their recombinant strains (Gupta et al. 2004; Hasan et al. 2006; Nagarajan 2012; Thakur 2012). The extracellular bacterial lipases are the main commercial lipases due to their easier bulk production. The major microbial producers for extracellular lipases are Mucor miehei, Rhizopus oryzae, Candida antarctica, and Pseudomonas cepacia, etc. (Gog et al. 2012; Gupta et al. 2015; Hasan et al. 2006; Joseph et al. 2008; Saxena et al. 2003). However, only a few bacterial are confirmed as sources for commercialization, such as Achromobacter, Alcaligenes, Arthrobacter, Bacillus, Burkholderia, Chromobacterium, and Pseudomonas. Of these, the lipases from Pseudomonas bacteria are preferable for use in a variety of biotechnological applications (Jaeger et al. 1994; Kapoor and Gupta 2012; Pandey et al. 1999).
Microbial lipases always present different structural and catalytic characteristics with respect to different sources. Many studies have determined the three-dimensional structures of many lipases by using X-ray crystallography, indicating that all the microbial lipases share a similar structure, such as α/β-hydrolase fold, which is composed of a core of predominantly parallel β strands surrounded by α helices. However, different microbial lipases show differences in structures. For instance, lipase B from Candida antarctica does not have a conserved pentapeptide sequence Gly-X-Ser-X-Gly around the active site, which is present in most of the other lipases (Gotor-Fernández et al. 2006; Gupta et al. 2015; Joseph et al. 2008; Uppenberg et al. 1994). The “lid” structure of lipase from Geobacillus thermocatenulatus has a complex structure in terms of a large percentage of the amino acids of the enzyme (Carrasco-Lopez et al. 2009; Kapoor et al. 2012). As concluded by Pleiss et al. (1998), lipases contain three subgroups on the basis of the geometry of the binding site: (i) lipases with a hydrophobic, crevice-like binding site located near the protein surface (lipases from Rhizomucor and Rhizopus); (ii) lipases with a funnel-like binding site (lipases from C. antarctica, Pseudomonas, and mammalian pancreas); and (iii) lipases with a tunnel-like binding site (lipase from Candida rugosa). Lipases isolated from different sources have a wide range of properties with respect to positional specificity (regiospecificity), fatty acid specificity, acyl migration, substrate specificity, stereospecificity, thermostability, pH optimum, etc. One could probably find a lipase from nature that would be suitable for the desired application. Applications of industrial enzymes allow the technologist to discover lipases that more closely approach the gentle, efficient processes in nature (Hasan et al. 2006; Verma et al. 2012).
The Burkholderia cepacia complex (Bcc) is a group that contains at least 17 closely related gram-negative bacteria species or genomovars with different biological properties. These bacteria are widely but heterogeneously distributed in the natural environment, such as soil, water, rhizospheres, plants, fungi, and animals, as well as hospital environments and infected humans (Vandamme and Dawyndt 2011; Vial et al. 2011). The Bcc is one of the most important lipase-producing bacterial genera, and lipases were identified in Bcc isolates as early as 1984 (McKevitt and Woods 1984). The lipase obtained from B. cepacia was the most studied due to its high tolerance towards organic solvents and aliphatic alcohol, superior thermal stability, and wide substrate specificity (Boran and Ugur 2016; Mathiazakan et al. 2016), B. cepacia lipases are widely applied enzymes in biotechnology, which especially show very high transesterification activity in organic solvents (Ungcharoenwiwat and H-Kittikun 2015).
Hundreds of studies can be found for Bcc-lipase production and characteristics; however, few of them consider the lipases produced by B. pyrrocinia. Therefore, investigation of lipase production from B. pyrrocinia will become profound and necessary research to expand the microbial resources for lipases producing. In the present study, a lipase producing strain B1213 screened from the soil was identified as B. pyrrocinia. As far as we know, this is the first to report lipase productivity of B. pyrrocinia. We hope some basic and interesting findings in this work would provide clues and ideas for the development of B. pyrrocinia lipases, such as genetic modification for catalytic properties promotion and deep application.
Materials and methods
Materials
A total of more than 200 soil samples were collected from fertile soils in Beijing, Shandong, Henan, Dalian, Sichuan, Hunan, Inner Mongolia, and some regions near the vegetable oil factory in China. A preliminary screening medium (natural pH) was used, and the composition (per liter) is listed as follows: agar, 18.0 g; emulsified olive oil (4% PVA: olive oil = 3:1, w/w), 120.0 mL; NaCl, 3.0 g; MgSO4·7H2O, 0.4 g; K2HPO4, 0.5 g; (NH4)2SO4, 0.5 g; rhodamine B (10 mg/mL), 1.0 mL. Submerged culture medium (per liter, natural pH) was prepared by saccharose, 5.0 g; soy peptone, 20.0 g; K2HPO4, 1.0 g; (NH4)2SO4, 1.0 g; MgSO4, 0.8 g; and olive oil 10.0 mL. Both media were sterilized at 115 °C for 30 min. Sephacryl S200 and Q-Sepharose™ Fast Flow (Q Sepharose FF) were purchased from General Electric Company (GE Health). Polyethylene glycol (PEG) 2000 was purchased from Sinopharm. p-nitrophenyl acetate (p-NPA), p-nitrophenyl benzoate (p-NPB), p-nitrophenyl caprylate (p-NPC), and p-nitrophenyl palmitate (p-NPP) were purchased from Shanghai yuanye biological technology co., LTD. All other the reagents were of at least analytical grade and were purchased from Beijing Aobox Biotechnology LLC (Beijing, China).
Screening, isolation and identification of lipase producing strains
In the preliminary screening process, soil suspensions were prepared by mixing 0.2 g soil samples into 1.0 mL of sterile water with vortex oscillation. Then, the suspensions with a concentration gradient of 10−5–10−3 were set and then inoculated onto the plate with preliminary screening medium using the spread-plate method. Each plate was cultured at 30 °C for 2–5 days.
In the rescreening process, colonies with rhodamine B color reaction were further screened and cultured using preliminary screening medium via the streak-plate method. Then the purified isolated strains were selected for further morphological and physiological characterization based on the analysis from the BLOLOG GEN III (Biolog, Inc., Hayward, CA, USA) and API 20E (BioMerieux, Shanghai, China) identification system. The molecular biological identification of isolated strains according to the 16S rDNA and recA sequence homology was reported by Beijing Haocheng Mingtai Technology CO., LTD. (Beijing, China), in which 27F 5′-AGAGTTTGATCCTGGCTCAG-3′ and 1492R 5′-ACG GTTACCTTGTTACGACTT-3′ were the 16S rDNA forward and reverse primers, respectively, and 5′-CTCTTCTTCGTCCATCGCCTC-3′ and 5′-TGACCGCCGAGAAGAGCAA-3′ were the BCR1 forward primer and BCR2 reverse primer for recA amplification, respectively.
Lipase activity assay
The activities of the lipase samples towards p-NPP were determined. The reaction mixtures contained 100 µL p-NPP (16.5 mM), 2.8 mL Tris–HCl buffer (0.2 M, 0.4% Triton X-100 and 0.1% Arabic gum, pH 8.0). After heating in the water bath for 5 min at 37 °C, 100 µL fermentation liquor was added, and heating was continued at the same temperature for another 10 min. The absorption value was obtained thereafter using a UV -spectrophotometer (TU-1900, Beijing Persee General Instrument Co. Ltd, Beijing, China) at 410 nm. One unit of lipolytic activity was defined as the amount of enzyme needed to release 1 mg of 4-nitrophenol per min under standard assay conditions.
Growth characteristics evaluation and lipase productivity optimization
The purified isolated strains from the rescreening process were transferred into Erlenmeyer flasks at pH 7.0, together with submerged culture medium containing 5.0 g of soy peptone, and the inoculum amount of strain was 1.0%. The 4 days-incubation was carried out in an orbital shaker (HZQ-Y100, Tiacang Laboratory equipment co., LTD., Suzhou, China) at 35 °C and 180 rpm. Then, 1 mL of fermentation liquor was taken out every 6 h to test the absorption value using a UV-spectrophotometer at 600 nm (TU-1900, Beijing Persee General Instrument Co. Ltd, Beijing, China). A time-OD value curve was recorded to reflect the growth characteristics of the new isolates. Meanwhile, fermentation liquor was also extracted every 6 h, and then centrifuged at 7000×g for 10 min (Allegra X-30R, Beckman Coulter, USA) to get the supernatant. A time-enzyme activity value curve was obtained by testing the lipase activity to reflect the lipase productivity along with the growth of the new isolates.
A one-factor experiment was carried out to evaluate the effect of different fermentation factors on the lipase productivity of the selected strain. Table 1 shows the detailed information of the factors and levels.
Table 1.
Fermentation factors and levels of one-factor experiment
| Factors | Types | Levels | Factors | Types | Levels |
|---|---|---|---|---|---|
| Carbon source | Sucrose | 5.0 g/L | Nitrogen source | Soy peptone | 20.0 g/L |
| Glucose | SP + (NH4)2SO4 | 20.0 + 1.0 g/L | |||
| Fructose | Beef peptone | 20.0 g/L | |||
| Lactose | BP + (NH4)2SO4 | 20.0 + 1.0 g/L | |||
| Maltose | Tryptone | 20.0 g/L | |||
| Oil | Olive oil | 10.0 mL/L | T + (NH4)2SO4 | 20.0 + 1.0 g/L | |
| Soybean oil | Urea | 20.0 g/L | |||
| Corn oil | U + (NH4)2SO4 | 20.0 + 1.0 g/L | |||
| Peanut oil | Surfactant | Tween-60 | 5.0 mL/L | ||
| Flaxseed oil | Tween-80 | ||||
| Triton X-100 |
| Factors | Levels | ||||
|---|---|---|---|---|---|
| Culture temperature | 25, 30, 35, 40, 45 °C | ||||
| Shaking speed | 120, 150, 180, 210, 240 r/min | ||||
| Inoculum amount | 2%, 4%, 6%, 8%, 10% | ||||
SP soy peptone, BP beef peptone, T tryptone, U urea
Lipase purification
An ordinal purification strategy that in the order of aqueous two-phase extraction (ATPE)–ion-exchange chromatography (IEC)–gel filtration chromatography (GFC) was adopted to purify the crude enzyme from the fermented liquid.
Aqueous two-phase systems containing PEG 2000 (5%), K2HPO4 (20%) and crude enzyme liquid (60%) were prepared at 25 °C. The mixture was centrifuged at 2000×g for the separation of the top and bottom phases. Then, the top phase was dialyzed (Membrane type: MD34mm: 10,000) and freeze dried. The enzyme powder was dissolved using Tris–HCl buffer solution (0.02 mM, pH 8.0) and filtered by a filter membrane (0.45 µm). Then, the filter liquor containing crude enzyme was further purified by IEC with a column (1 cm × 10 cm) filled with Q Sepharose Fast Flow, under the working conditions of: Tris–HCl buffer solution as a balance buffer solution (50 mM, pH 6.0), Tris–HCl buffer solution (50 mM, pH 6.0) with NaCl (1 M) as the elution buffer solution; the flow rate was 1 mL/min at 25 °C. The eluted enzymes were monitored using a diode array detector at 280 nm to select the secondary purified enzyme solution for further GFC purification. A Sephacryl S200 (1.5 cm × 50 cm) GFC column was used. The column was previously equilibrated with 0.2 M sodium phosphate buffer (pH 7.5). The enzyme was eluted with same buffer at flow rate of 0.8 mL/min at 25 °C, and the eluted enzyme was monitored using diode array detector at 280 nm. Both IEC and CFG were carried out using a chromatography system (AKTA 25, GE Health).
Polyacrylamide gel-electrophoresis analysis
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) was carried out to measure the molecular weight of the purified lipase from GFC. The concentrations of the separating and stacking gel were 12.5% and 4.5%, respectively. The lipase sample and sample buffer were mixed in a volume ratio of 3:1 and were kept in a boiling water bath for 5 min. The supernatant was injected into the gel lane after the mixture was centrifuged for 5 min at a speed of 1000×g. Electrophoresis was performed at a constant voltage of 80 V for approximately 1–2 h. The gel was stained with a Coomassie Brilliant Blue R250 of 0.25% (w/v), 50% (v/v) methanol and 10% (v/v) acetic acid. The gel was destained using the buffer with 25% (v/v) methanol and 7% (v/v) acetic acid.
Biochemical characterization evaluation of purified B. pyrrocinia lipase
pH and temperature tolerance
The optimal pH was determined by assaying the purified enzyme at different pH (6.0–11.0) using the following buffer systems: glycine–NaOH buffer (pH 9.0–11.0), Tris–HCl buffer (pH 7.0–9.0), sodium phosphate buffer (pH 6.0–7.5), acetic acid/sodium acetate buffer (4.5–5.5), and glycine–HCl buffer (pH 3.0–4.0). The pH stability of the enzyme was determined after incubation of the enzyme (1 U/mg) in the respective buffers (20 mM) of varying pH (3.0–11.0) for 12 h at 4 °C. The residual activity was measured under standard enzyme test conditions.
The optimum temperature of the purified lipase was determined by assaying at different temperatures (30–80 °C) using p-NPP as the substrates, and Tris–HCl buffer (pH 8.0) as buffer solution. The reaction mixture (in absence of lipase) was pre-heated to the desired temperature for 5 min in a water bath. Lipolysis was initiated by adding the enzyme (1 U/mg), and the activity was measured after a 10-min reaction. Thermal stability of the enzyme was estimated by incubating the enzyme at different temperatures (40–70 °C) for different times (10–60 min). The residual activity was measured under standard assay conditions.
Organic solvents and metal ions tolerance
The effect of organic solvents on lipase stability was determined by incubating the purified lipase in the presence of various organic solvents (75%, v/v). The reactions were incubated in the water bath at 30 °C for 10 min. The stability is expressed as the residual lipolytic activity relative to the enzyme control incubated without organic solvent.
The effect of the metal ions on lipase stability was determined by incubating the purified lipase in the presence of metal ions (Cu2+, Fe3+, Fe2+, Ca2+, K+, Zn2+, Mg2+, Mn2+, Al3+) at a concentration of 1 mM and 5 mM (final concentration) prepared by Tris–HCl buffer (50 mM, pH 8.0). The reactions were incubated in the water bath at 30 °C for 10 min. The relative activity of the enzyme was calculated by comparison with enzyme incubated under similar conditions without metal salts (As blank group, relative activity = 100%).
Substrate specificity and reaction kinetic
The substrate specificity of the lipase was investigated using 4-nitrophenyl esters with different acyl chain lengths (p-NPA (C2), p-NPB (C4), p-NPC (C8), P-NPP (C16), 16.5 mM), and lipolytic activity was measured under standard assay conditions mentioned as Lipase activity assay section, by replacing P-NPP with each of the above four 4-nitrophenyl esters separately. To evaluate the reaction kinetics of the lipase, p-NPP (C16) was used as the substrate at different concentration. The Michaelis–Menten constant (Km) and the maximum specific activity (Vmax) were calculated from the Lineweaver–Burk plots using Origin 8.0 software.
Statistical analysis
All experiments were performed at least twice using freshly prepared samples. The mean and standard deviations calculated from these measurements were plotted. Origin (Origin Lab Co., Pro. 8.0) was used for processing the data and creating charts. An analysis of variance (ANOVA) was performed to examine the differences between the means using Tukey’s post hoc test and using SPSS at α level of 0.05. Design-Expert was used to build a regression fitting equation and analyze the significance.
Results and discussion
Isolation and identification of lipase producing microorganism
In this study, over 200 soil samples were collected as the source of lipase-producing microorganisms, of which 42 strains were selected for re-culture in a liquid culture medium due to their capacity to grow on the plate medium with rhodamine B, which was accompanied by a color circle phenomenon (Fig. S1a). In the experiment, parts of the selected strains have lipase activities, and strain B1213 showed the most significant lipase activity of approximately 70.73 U/mL, which was much higher than the others. Hence, this B1213 was physiologically and biochemically identified.
After observing the growth of B1213 on the solid medium and the morphology of B1213 under a microscope, it was found that the B1213 was a short rod-shaped gram-negative bacterium (Fig. S1b) with round, light yellow and opaque colonies on solid medium (Fig. S1c).
The 16S rDNA and recA of B1213 were sequenced and compared to other organisms in the NCBI database by BLAST analysis. The inter-comparison results suggested a close relationship between strain B1213 and the members of the Burkholderia cepacia species, with a maximum sequence homology (99%) to B. stabilis and B. pyrrocinia. According to the phylogenetic trees (Fig. S2) based on the sequences of the 16S rDNA and recA, as well as the physiological and biochemical analysis, strain B1213 was identified as B. pyrrocinia and has been deposited in the China General Microbiological Culture Collection Center (CGMCC, No.: 12806).
Growth and lipase producing characteristics of B1213
It is obvious that there is a high correlation between cell metabolism and enzyme production. Therefore, confirming the metabolic characteristics of B1213 is the basis for selecting the correct strain with high lipase productivity and a high metabolic rate. As shown in Fig. 1a, the dynamic growth level reflected by the cell amount in the fermentation liquor was detected using a turbidity test. In the same period, the dynamic lipase productivity was also recorded. It was found that B1213 began its logarithmic phase very quickly after culture fermentation for 2 h and lasted nearly 18 h. Nonetheless, the B1213 turned into stationary phase from the 20th to 72nd hours. Figure 1a also showed the ability of lipase production increased in all the phases, reaching a peak at the 72nd hour. Afterwards, the growth rate declined, resulting from the low metabolism level of B1213 in the aging phase. Therefore, further fermentation optimization conditions and flask shaking culture was conducted from the seed of living B1213 at the 72nd hour.
Fig. 1.

Growth and lipase productivity curve (a) and the effect of medium composition, such as the carbon source (b), nitrogen source (c), oil (d), and surfactant €, on the lipase activity of B1213. SP soy peptone, BP beef peptone, T tryptone, U urea
Optimized conditions for lipase production
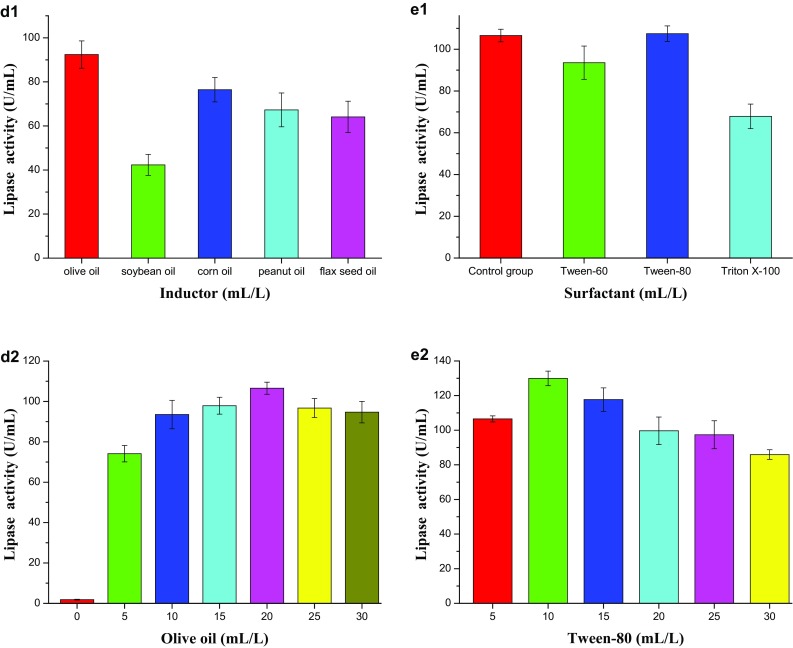
Microbial fermentation is a complex system affected by many factors, such as the inoculum amount, nutrition and fermentation conditions. This study investigated the single influence of different factors. When considering the effect of the carbon source on lipase production (Fig. 1b-1), sucrose appears to be the most favorable source for lipase production of B1213, whereas glucose occurs as the most unfavorable one. As a result, sucrose was selected as the carbon source, and its impact on the different additions was investigated. As show in Fig. 1b-2, the addition of sucrose had a significant effect on the production of lipase. When the concentration of sucrose was 1.0 g/L, the highest yield of approximately 77 U/mL (Approx.) was obtained. Hence, 1.0 g/L of sucrose was suggested as the optimal addition of carbon source.
When considering the effect of the nitrogen source on lipase production, both organic (20.0 g/L) and mixed nitrogen (20.0 + 1.0 g/L) were investigated. The results (Fig. 1c-1) showed that only the beef peptone played a similar role to the mixed peptone that was combined with beef peptone and the (NH4)2SO4. But the other mixed nitrogen sources were little slightly better than the organic one in lipase production. It is clear that the mixed nitrogen source that contained 20.0 g/L tryptone and 1.0 g/L (NH4)2SO4 shows the best effect on lipase yield, followed by the organic 20.0 g/L tryptone nitrogen source. The ascertained results clearly demonstrate that B1213 could not thrive well when urea were used as nitrogen source. Due to the similar influence of tryptone and tryptone plus (NH4)2SO4, the tryptone was selected to evaluate its influence on lipase production under different addition. Results showed that the tryptone concentration in the culture medium had a great effect on lipase production from B1213. When the concentration of tryptone was increased up to 25.0 g/L (Fig. 1c-2), the yield of enzyme also increased directly, suggesting that the correct amount of tryptone needed for enzyme activity was 25.0 g/L.
The lipids used in this system are not just carbon source in the synthesis, but they also play the role of inducers for lipase production. Our assessment outcome from Fig. 1d-1, d-2 indicated that there was no lipase produced by B1213 in the fermentation liquid absence of oil. Conversely, olive oil causeed high lipase production rate, followed by corn oil and soybean. An increment of olive oil concentration of 20.0 mL/L increased the activity of lipase production to a peak of 106.5 U/mL and later began to decrease slightly, owing to the possible reason that high concentration of oil might inhibit or hinder the enzymatic activities.
Surfactants can emulsify oil, making a better mixture between the oil and medium, and increase the contact area between the cell and oil. Additionally, the surfactants can also change the permeability of the cell membrane and improve the transmission speed of oxygen at the gas–liquid interface. As shown in the Fig. 1e-1, e-2, at the level of 5.0 mL/L, the addition of Tween-60 and Triton X-100 did not improve, but inhibit the lipase yield, indicating that the strain B1213 has a certain selectivity to the surfactant. The addition of 10.0 mL/L Tween-80 helped to increase the production of lipase to a maximum peak level. However, the lipase activity decreased negatively with an upward increment of Tween-80. Conclusively, the optimal amount of Tween-80 was 10.0 mL/L.
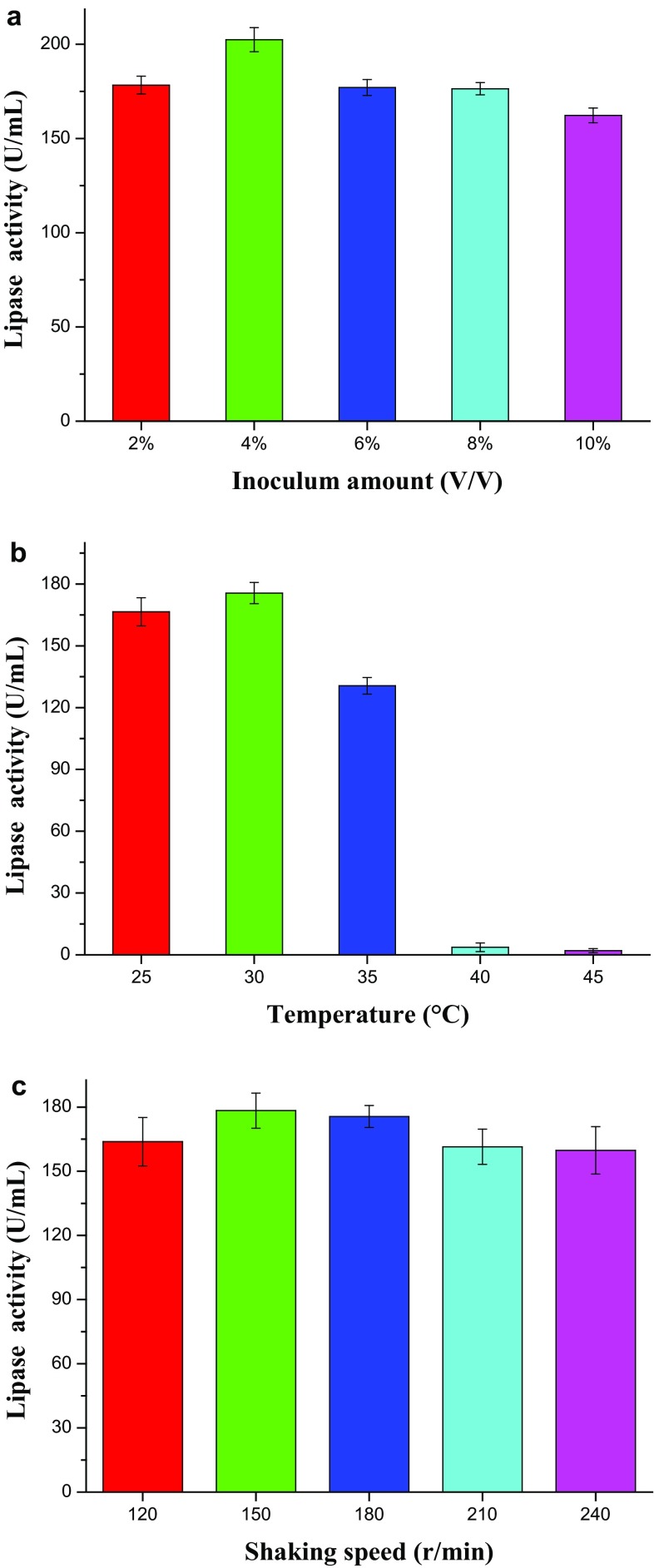
On the other hand, fermentation conditions, including inoculation rates, temperature, and shaking speed, are other factors that can affect the fermentation cycle and the level of enzyme production. Lower inoculation will lead to lower cell viability, which is not conducive to enzyme production. Larger inoculation will bring more cultivable metabolites, leading strain to the ease of early aging. Only the appropriate inoculation amount is helpful to obtain enough cells in a short time for enzyme production. The results (Fig. 2a) showed that when the inoculum amount was 4%, the strain B1213 could produce the lipase at the highest efficiency.
Fig. 2.
Effect of fermentation conditions, such as inoculum size (a), temperature (b), and shaking speed (c), on the lipase activity of B1213
The current study of fermentation was carried out at a temperature from 25 to 45 °C. The results shown in Fig. 2b indicated that the optimum temperature for B1213 to produce lipase was 30 °C, and when the temperature was higher than 40 °C, the lipase productivity was greatly inhibited. The above results also showed that the lipase from B1213 might belong to mesophilic enzyme, which is sensitive to the higher temperature. When considering the influence of shaking speed, the results in Fig. 2c indicated that the influence was not very significant, which means that the oxygen demand of B1213 is small for lipase production.
Purification of B. pyrrocinia lipase
The extracellular lipase from B. pyrrocinia B1213 was purified by a three-step purification process followed of ATPE-IEC-GFC. As the purification results summarized (see Table S1), the lipase from B1213 was obtained a 11.1-fold increase in specific activity, with 10.3% recovery of overall yield. The final specific activity of purified lipase reached 875.7 U/mg protein. Though the purification efficiency was high to obtain high-quality purified lipase, the research experience in this study indicated that the purification process of IEC might be a critical step to affect the total yield, which means there will be the potential to promote the yield in the IEC step. As reported by Castro-Ochoa et al.(2005; Sugimura et al. 2000), many microbial lipases, especially lipases from Burkholderia (bas Pseudomonas) cepacian (Salameh and Wiegel 2010) perform self-aggregation in solution, which could be due to the high content of hydrophobic amino acids and surface hydrophobic characteristics of lipases. The low yield of the lipase in purification could be due to the aggregation of the protein (Lesuisse et al. 1993; Velu et al. 2012).
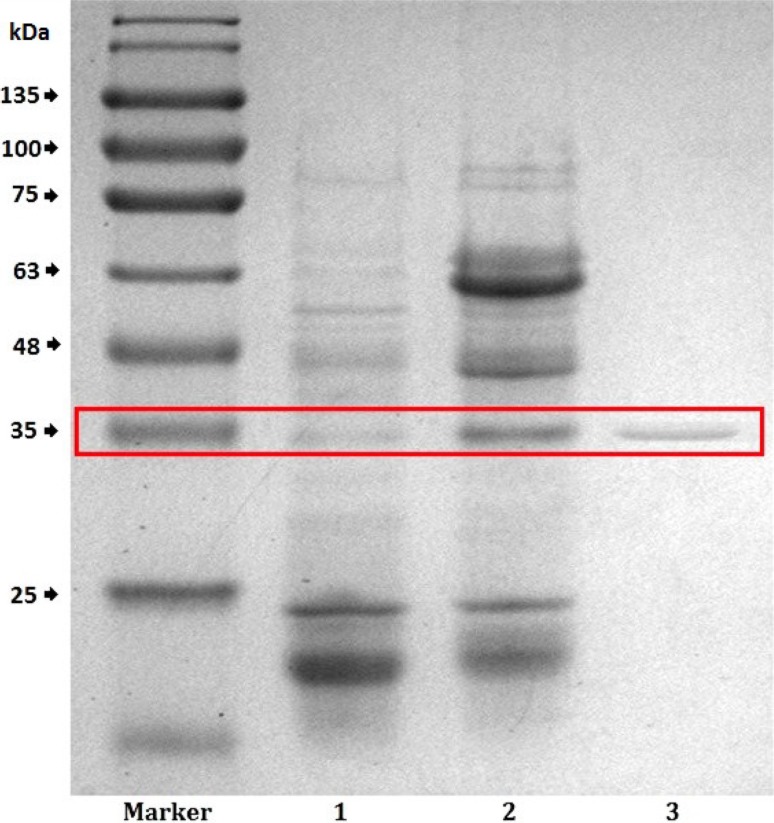
To avoid the negative impact from lipase aggregation, denaturing PAGE analysis was followed (Fig. 3), and the molecular weight of the purified lipase was calculated to be 35 kDa.
Fig. 3.
SDS–PAGE electrophoretogram of lipase from different purification strategies. Lane 1: crude enzyme, Lane 2: purified enzyme from ATPE, Lane 3: final purified enzyme from GFC
pH and temperature tolerance of lipase
The effect of the pH on lipase activity and stability was shown in Fig. 4a. In the pH range from 6.0 to 11.0, the optimum pH of this lipase was found to be 8.0. A sharp increase in activity was observed from pH 6.0 (15.5%) to 8.0 (100%), and the relative activity decreased to 61.9% when the pH increased to 10.5. Meanwhile, after 12 h of storage at 4 °C, the lipase remained relatively stable in the pH range 6.0–9.0, indicating that this lipase from B1213 belongs to alkaline lipases, similar to most of other lipases from B. pyrrocinia.
Fig. 4.

Effect of pH (a), temperature (b, c), organic solvents (d) and metal ions (e) on activity and stability of lipase
The result from Fig. 4b indicated that this lipase was found to be active in the temperature range of 30–80 °C, and the optimum temperature was 50 °C, whereas approximately 78% and 59% activity was observed at 40 and 60 °C, respectively. When considering the thermal stability of the lipase after incubating the lipase at different temperatures (40–70 °C), the results clearly indicated that the increase in the incubation temperature and time enhanced the negative impact on lipase stability. After 1 h, the enzyme retained 88% activity at 40 °C, 36% activity at 50 °C and nearly complete inactivation at 70 °C (Fig. 4c), which demonstrated that the current lipase was not suitable for application at high temperature.
Organic solvents and metal ions tolerance of lipase
The activity of the enzymes is strongly affected by different solvent. Based on the widely accepted model for explaining the solvent effects (Laane et al. 1987), the enzyme activity is higher in the environment surrounded by nonpolar and mid polar solvents, whereas it is the lowest in the polar solvents. The polarity of the solvent used in our experiments is in the following order: methanol > ethanol > acetone > isopropanol > n-hexane. The effects of these organic solvents on the stability of lipase B1213 are shown in Fig. 4d. Compared to the control sample, isopropanol showed the smallest influence, followed by acetone, and both retained more than 85% enzyme activity at 30 °C for 10 min. However, contrary to Laane’s suggestion model, n-hexane would reduce lipase activity to 10.6%, which was the most negative organic solvent.
As shown in Fig. 4e, Mg2+, Al3+, Mn2+ and Fe3+ could enhance the lipase activity at both concentrations. Except Ca2+, Fe3+ and Fe2+, all other ions we investigated showed a negative effect when the concentration was increased form 1 to 5 mM, especially Zn2+.
Substrate specificity and reaction kinetics
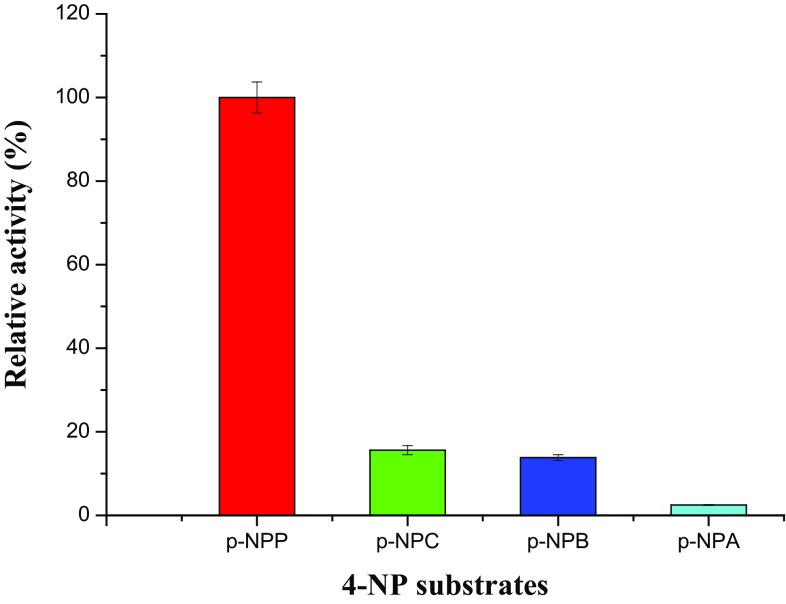
Four 4-NPP substrates of varying acyl chain length were used to test the chain length specificity of lipase B1213. The results shown in Fig. 5, the lipase B1213 showed a very narrow range of substrate chain length specificity. Long-chain acyl group 4-NP esters (p-NPP) seemed to be good substrates, while nearly no reaction happened with the short-chain acyl group 4-NP esters (p-NPA). Our results also indicated that, when p-NPP was a substrate, the Michaelis constant, Km of lipase B1213 was 0.273 mmol/L, and the maximal velocity (Vmax) was 909.09 U/mg.
Fig. 5.
Acyl chain length specificity of the lipase against 4-NP substrates
Conclusions
Microbial lipases from different sources always present different structural and catalytic characteristics. This study was the first report referred to lipase productivity of B. pyrrocinia, which will not only be meaningful for resource expansions of lipase-producing microorganisms but also to shed light on exploring the characteristics of lipases from B. pyrrocinia. In the present study, tryptone, olive oil and temperature had significant effects on lipase production. Under the single-factor and response surface optimization process, the final lipase yield was up to 205.1 U/mL, making this B1213 a potential high-yield strain for lipases. Thus, this B1213 will be valuable in doing further research on lipase productivity. Based on ATPE-IEC-GFC purification strategy, the specific activity of lipase reached 75.7 U/mg protein, with a molecular weight of 35 kDa. We suggest paying more attention to the IEC step to promote the yield of the purification.
Our results show that this lipase from B1213 was one kind of alkaline lipase. The optimum pH and temperature of this lipase was pH 8.0 and 50 °C respectively, and nearly completely inactivated at 70 °C, which demonstrates that the lipase is not suitable for application in areas of high temperature. However, it also gave us information that this B1213 lipase can exert the best catalytic ability in a mild environment, making catalytic reactions easy to achieve.
Except the application potentiality, we suggest deep research should still be performed to explore the characteristics of lipase B1213, especially its molecular structure, catalytic properties, genetic modification.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Numbers: 31501487, 31671798). We are grateful to Professor Madhav P. Yadav for his helpful promoting in the language quality of this article.
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Aguieiras ECG, Cavalcanti-Oliveira ED, Freire DMG. Current status and new developments of biodiesel production using fungal lipases. Fuel. 2015;159:52–67. doi: 10.1016/j.fuel.2015.06.064. [DOI] [Google Scholar]
- Boran R, Ugur A. Burkholderia multivorans SB6 lipase as a detergent ingredient: characterization and stabilization. J Surfactants Deterg. 2016;19(1):39–48. doi: 10.1007/s11743-015-1767-6. [DOI] [Google Scholar]
- Carrasco-Lopez C, Godoy C, de Las RB, Fernandez-Lorente G, Palomo JM, Guisan JM, Fernandez-Lafuente R, Martinez-Ripoll M, Hermoso JA. Activation of bacterial thermoalkalophilic lipases is spurred by dramatic structural rearrangements. J Biol Chem. 2009;284(7):4365–4372. doi: 10.1074/jbc.M808268200. [DOI] [PubMed] [Google Scholar]
- Castro-Ochoa LD, Rodríguez-Gómez C, Valerio-Alfaro G, Ros O. Screening, purification and characterization of the thermoalkalophilic lipase produced by Bacillus thermoleovorans CCR11. Enzyme Microbial Technol. 2005;37(6):648–654. doi: 10.1016/j.enzmictec.2005.06.003. [DOI] [Google Scholar]
- de Abreu L, Fernandez-Lafuente R, Rodrigues RC, Volpato G, Ayub MAZ. Efficient purification-immobilization of an organic solvent-tolerant lipase from Staphylococcus warneri EX17 on porous styrene-divinylbenzene beads. J Mol Catal B: Enzym. 2014;99:51–55. doi: 10.1016/j.molcatb.2013.10.018. [DOI] [Google Scholar]
- Feng X, Patterson DA, Balaban M, Emanuelsson EAC. Characterization of tributyrin hydrolysis by immobilized lipase on woolen cloth using conventional batch and novel spinning cloth disc reactors. Chem Eng Res Des. 2013;91(9):1684–1692. doi: 10.1016/j.cherd.2013.06.009. [DOI] [Google Scholar]
- Gog A, Roman M, Toşa M, Paizs C, Irimie FD. Biodiesel production using enzymatic transesterification – Current state and perspectives. Renewable Energy. 2012;39(1):10–16. doi: 10.1016/j.renene.2011.08.007. [DOI] [Google Scholar]
- Gotor-Fernández V, Busto E, Gotor V. Candida antarctica lipase B: an ideal biocatalyst for the preparation of nitrogenated organic compounds. Adv Synth Catal. 2006;348(7–8):797–812. doi: 10.1002/adsc.200606057. [DOI] [Google Scholar]
- Gupta R, Gupta N, Rathi P. Bacterial lipases: an overview of production, purification and biochemical properties. Appl Microbiol Biotechnol. 2004;64(6):763–781. doi: 10.1007/s00253-004-1568-8. [DOI] [PubMed] [Google Scholar]
- Gupta R, Kumari A, Syal P, Singh Y. Molecular and functional diversity of yeast and fungal lipases: their role in biotechnology and cellular physiology. Prog Lipid Res. 2015;57:40–54. doi: 10.1016/j.plipres.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Gutarra MLE, Godoy MG, Maugeri F, Rodrigues MI, Freire DMG, Castilho LR. Production of an acidic and thermostable lipase of the mesophilic fungus Penicillium simplicissimum by solid-state fermentation. Biores Technol. 2009;100(21):5249–5254. doi: 10.1016/j.biortech.2008.08.050. [DOI] [PubMed] [Google Scholar]
- Hasan F, Shah AA, Hameed A. Industrial applications of microbial lipases. Enzym Microbial Technol. 2006;39(2):235–251. doi: 10.1016/j.enzmictec.2005.10.016. [DOI] [Google Scholar]
- Jaeger KE, Ransac S, Dijkstra BW, Colson C, van Heuvel M, Misset O. Bacterial lipases. FEMS Microbiol Rev. 1994;15(1):29. doi: 10.1111/j.1574-6976.1994.tb00121.x. [DOI] [PubMed] [Google Scholar]
- Joseph B, Ramteke PW, Thomas G. Cold active microbial lipases: Some hot issues and recent developments. Biotechnol Adv. 2008;26(5):457–470. doi: 10.1016/j.biotechadv.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Joseph B, Upadhyaya S, Ramteke P. Production of cold-active bacterial lipases through semisolid state fermentation using oil cakes. Enzym Res. 2011;2011:16. doi: 10.4061/2011/796407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Gupta MN. Lipase promiscuity and its biochemical applications. Process Biochem. 2012;47(4):555–569. doi: 10.1016/j.procbio.2012.01.011. [DOI] [Google Scholar]
- Laane C, Boeren S, Vos K, Veeger C. Rules for optimization of biocatalysis in organic solvents. Biotechnol Bioeng. 1987;30(1):81–87. doi: 10.1002/bit.260300112. [DOI] [PubMed] [Google Scholar]
- Lesuisse E, Schanck K, Colson C. Purification and preliminary characterization of the extracellular lipase of Bacillus subtilis 168, an extremely basic pH-tolerant enzyme. Eur J Biochem. 1993;216(1):155–160. doi: 10.1111/j.1432-1033.1993.tb18127.x. [DOI] [PubMed] [Google Scholar]
- Maiangwa J, Ali MSM, Salleh AB, Rahman RNZR, Shariff FM, Leow TC. Adaptational properties and applications of cold-active lipases from psychrophilic bacteria. Extremophiles. 2015;19(2):235–247. doi: 10.1007/s00792-014-0710-5. [DOI] [PubMed] [Google Scholar]
- Masomian M, Rahman RNZR, Salleh AB, Basri M. A new thermostable and organic solvent-tolerant lipase from Aneurinibacillus thermoaerophilus strain HZ. Process Biochem. 2013;48(1):169–175. doi: 10.1016/j.procbio.2012.11.002. [DOI] [Google Scholar]
- Mathiazakan P, Shing SY, Ying SS, Kek HK, Tang MSY, Show PL, Ooi C, Ling TC. Pilot-scale aqueous two-phase floatation for direct recovery of lipase derived from Burkholderia cepacia strain ST8. Sep Purif Technol. 2016;171:206–213. doi: 10.1016/j.seppur.2016.07.017. [DOI] [Google Scholar]
- McKevitt AI, Woods DE. Characterization of pseudomonas cepacia isolates from patients with cystic fibrosis. J Clin Microbiol. 1984;19(2):291–293. doi: 10.1128/jcm.19.2.291-293.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan S. New tools for exploring “Old friends—microbial lipases”. Appl Biochem Biotechnol. 2012;168(5):1163–1196. doi: 10.1007/s12010-012-9849-7. [DOI] [PubMed] [Google Scholar]
- Pandey A, Benjamin S, Soccol CR, Nigam P, Krieger N, Soccol VT. The realm of microbial lipases in biotechnology. Biotechnol Appl Biochem. 1999;29(Pt 2):119–131. [PubMed] [Google Scholar]
- Pleiss J, Fischer M, Schmid RD. Anatomy of lipase binding sites: the scissile fatty acid binding site. Chem Phys Lipid. 1998;93(1–2):67–80. doi: 10.1016/S0009-3084(98)00030-9. [DOI] [PubMed] [Google Scholar]
- Salameh MA, Wiegel J. Effects of detergents on activity, thermostability and aggregation of two alkalithermophilic lipases from thermosyntropha lipolytica. Open Biochem. 2010;4(2):22–28. doi: 10.2174/1874091X01004010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salihu A, Alam MZ. Solvent tolerant lipases: a review. Process Biochem. 2015;50(1):86–96. doi: 10.1016/j.procbio.2014.10.019. [DOI] [Google Scholar]
- Saxena RK, Sheoran A, Giri B, Davidson WS. Purification strategies for microbial lipases. J Microbiol Methods. 2003;52(1):1–18. doi: 10.1016/S0167-7012(02)00161-6. [DOI] [PubMed] [Google Scholar]
- Sugimura Y, Fukunaga K, Matsuno T, Nakao K, Goto M, Nakashio F. A study on the surface hydrophobicity of lipases. Biochem Eng J. 2000;5(2):123–128. doi: 10.1016/S1369-703X(99)00072-8. [DOI] [PubMed] [Google Scholar]
- Thakur S. Lipases, its sources, properties and applications: a review. Int J Sci Eng Res. 2012;3(7):1–29. [Google Scholar]
- Ungcharoenwiwat P, H-Kittikun A. Purification and characterization of lipase from Burkholderia sp. EQ3 isolated from wastewater from a canned fish factory and its application for the synthesis of wax esters. J Mol Catal B Enzym. 2015;115:96–104. doi: 10.1016/j.molcatb.2015.02.005. [DOI] [Google Scholar]
- Uppenberg J, Hansen MT, Patkar S, Jones TA. The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica. Structure. 1994;2(4):293–308. doi: 10.1016/S0969-2126(00)00031-9. [DOI] [PubMed] [Google Scholar]
- Vandamme P, Dawyndt P. Classification and identification of the Burkholderia cepacia complex: past, present and future. Syst Appl Microbiol. 2011;34(2):87–95. doi: 10.1016/j.syapm.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Velu N, Divakar K, Nandhinidevi G, Gautam P. Lipase from Aeromonas caviae AU04: isolation, purification and protein aggregation. Biocatal Agric Biotechnol. 2012;1(1):45–50. [Google Scholar]
- Verma N, Thakur S, Bhatt AK. Microbial lipases: industrial applications and properties (a review) Int Res J Biol Sci. 2012;1(8):88–92. [Google Scholar]
- Vial L, Chapalain A, Groleau M, Déziel E. The various lifestyles of the Burkholderia cepacia complex species: a tribute to adaptation. Environ Microbiol. 2011;13(1):1–12. doi: 10.1111/j.1462-2920.2010.02343.x. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhang K, Huang Y, Wang Z. Both hydrolytic and transesterification activities of Penicillium expansum lipase are significantly enhanced in ionic liquid [BMIm][PF6] J Mol Catal B Enzym. 2010;63(1–2):23–30. doi: 10.1016/j.molcatb.2009.11.014. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.