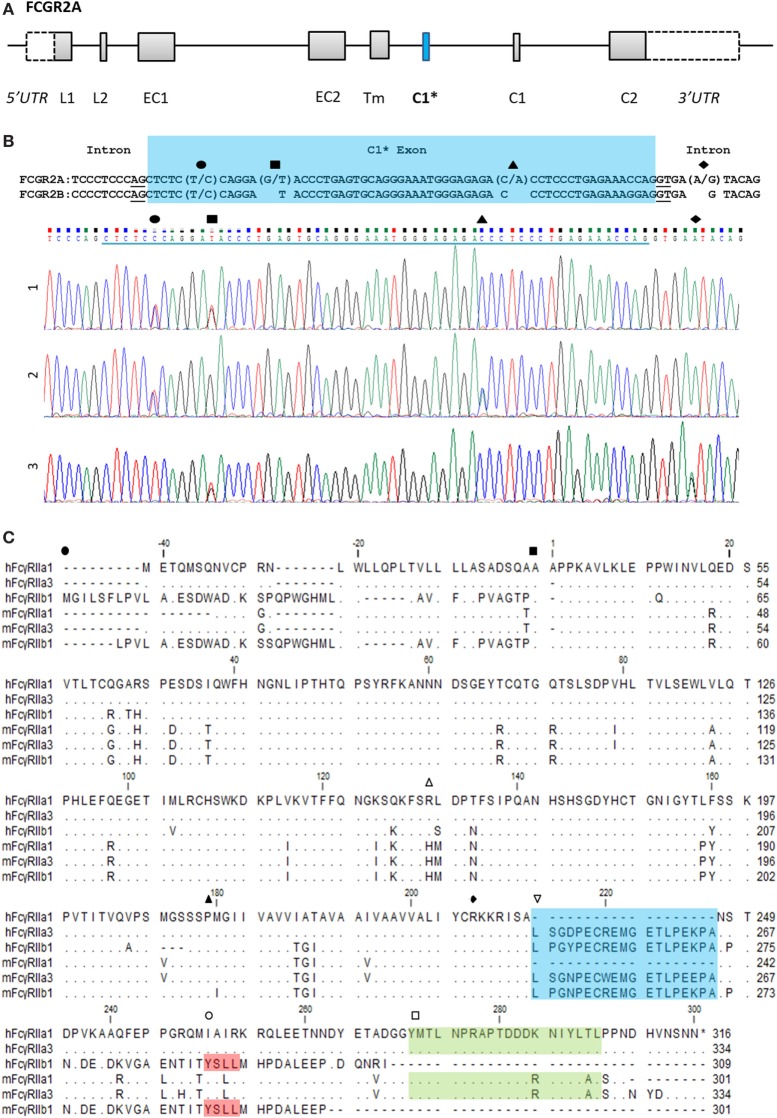
Figure 1.
FCGR2A organization and the C1* exon. (A) Organization of the FCGR2A gene. Exonic regions are represented as boxes and intronic regions as a line. Leader, (EC) extracellular, (Tm) transmembrane, and (C) cytoplasmic tail. (B) Alignment of genomic DNA sequence surrounding the C1* exon of FCGR2A and FCGR2B with single nucleotide polymorphisms and exemplar electropherograms of FcγRIIa3 from three individuals. SNPs are indicated thus: exonic (●) C/T, (■) G/T, and (▲) C/A SNPs which result in proline to serine, aspartic acid to tyrosine, or threonine to asparagine substitution, respectively. The intronic A/G SNP, (◆), in electropherogram 3, reported to promote splicing of C1* exonic sequences results in a unique 19-amino acid insertion in the FcγRIIa3 cytoplasmic tail. (C) Amino acid alignment of human (h) and macaque (m) FcγRIIa1, FcγRIIa3, and FcγRIIb1 (32). cDNA sequences were derived from healthy human or macaque PBMC samples. Dot (∙) represents residues matched to human FcγRIIa1 sequence; dash (-) represents a gap in sequence alignment. The N-terminal end of different FcγR structural domains are indicated: (●) leader sequence, (■) extracellular domain, (▲) transmembrane region, and (◆) cytoplasmic tail. Regions of interest are denoted with ( ) polymorphic 131 residue, (
) polymorphic 131 residue, ( ) C1 exon (blue), (
) C1 exon (blue), ( ) ITIM in FcγRIIb sequence (red), and (
) ITIM in FcγRIIb sequence (red), and ( ) immunoreceptor tyrosine activation motif FcγRIIa (green) regions highlighted.
) immunoreceptor tyrosine activation motif FcγRIIa (green) regions highlighted.