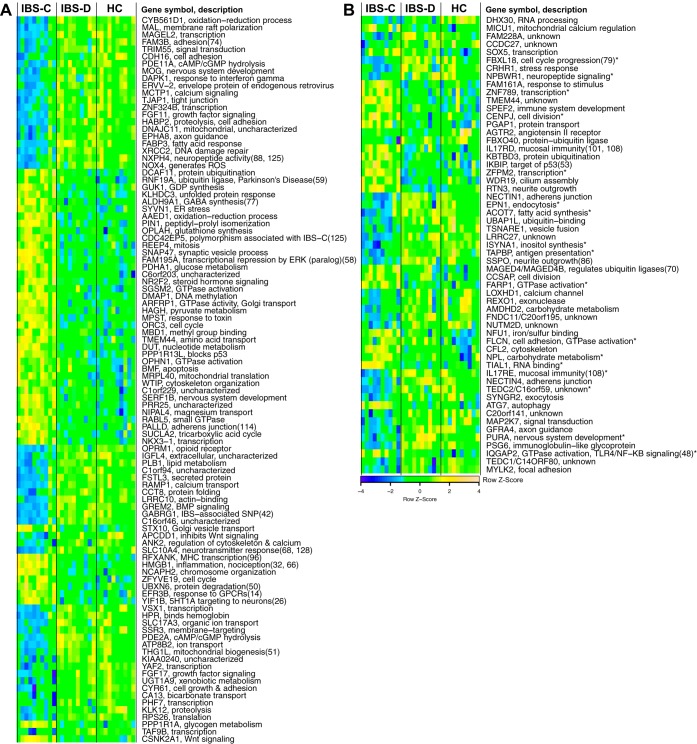
Fig. 1.
A: top 100 differentially expressed genes in irritable bowel syndrome with constipation (IBS-C) vs. healthy controls (HCs). B: differentially expressed genes in IBS-C vs. HC that are replicated in the Mayo Clinic cohort. Expression values are scaled. Descriptions are from gene ontology or KEGG pathways, except where referenced. Genes and samples (within bowel habit subtypes) are clustered by Euclidean distance. Sample clustering is based on top 500 probes after ordering by variance across all 30 sample. IBS-D, IBS with diarrhea; cAMP/cGMP, cyclic adenosine/guansosine monophosphate; GTP, guanosine triphosphate; GTPase, enzymatic function catalyzing formation of GDP from GTP; BMP, bone-morphogenic protein; SNP, single nucleotide polymorphism; MHC, major histocompatibility complex; ATP, adenosine triphosphate; GPCR, G protein-coupled receptor; PKA, protein kinase A.