Abstract
The cellular prion protein (PrPC) can act as a cell-surface receptor for β-amyloid (Aβ) peptide; however, a role for PrPC in the pathogenesis of Alzheimer's disease (AD) is contested. Here, we expressed a range of Aβ isoforms and PrPC in the Drosophila brain. We found that co-expression of Aβ and PrPC significantly reduces the lifespan, disrupts circadian rhythms, and increases Aβ deposition in the fly brain. In contrast, under the same conditions, expression of Aβ or PrPC individually did not lead to these phenotypic changes. In vitro studies revealed that substoichiometric amounts of PrPC trap Aβ as oligomeric assemblies and fragment-preformed Aβ fibers. The ability of membrane-anchored PrPC to trap Aβ as cytotoxic oligomers at the membrane surface and fragment inert Aβ fibers suggests a mechanism by which PrPC exacerbates Aβ deposition and pathogenic phenotypes in the fly, supporting a role for PrPC in AD. This study provides a second animal model linking PrPC expression with Aβ toxicity and supports a role for PrPC in AD pathogenesis. Blocking the interaction of Aβ and PrPC represents a potential therapeutic strategy.
Keywords: Alzheimer's disease, Drosophila, prion, amyloid-beta (AB), oligomer, protein misfolding, fibril, neurodegenerative disease, animal model, circadian rhythm
Introduction
Alzheimer's disease (AD)3 is the most prevalent adult neurodegenerative disease. It affects 46 million people worldwide, and this number is predicted to almost triple by 2050 (1). There is abundant evidence that self-associated assemblies of β-amyloid peptides (Aβ) have a crucial role in initiating the pathological cascade that culminates in neuronal dysfunction and death. A particularly toxic activity has been assigned to low molecular weight, soluble oligomers of Aβ, rather than amyloid fibrils (2–5). The pathological mechanisms are unclear but may include toxic interactions of the oligomers with cellular membranes, perhaps inducing ion channel formation (6). In addition, a number of more-or-less specific interactions of oligomeric Aβ with putative receptor proteins have been described (7, 8). A seminal study by Laurén et al. (9) focused attention on the prion protein (PrP) as the only high-affinity binder of Aβ oligomers found in a 200,000-strong human cDNA library.
The N terminus of PrPC binds to Aβ oligomers with nanomolar affinity (9–14), and in some model systems this interaction appears to be essential for synapto- and neurotoxicity. Specifically, Gimbel et al. (13) found that transgenic mice used as a model of Aβ toxicity were rendered resistant to pathology if the PrP gene was knocked out. This rescue from the damaging effects of Aβ was observed despite the amyloid plaque density and glial responses being essentially identical to control PrP+/+ mice. Data indicating that PrP binds to assemblies of oligomeric Aβ in human brains (15) and that extracts from AD brain extracts require PrP to suppress hippocampal long term potentiation (10) lend support to the importance of the PrP–Aβ interaction in AD pathogenesis (8). PrPC has also been shown to exacerbate Aβ impairment of synaptic plasticity (12). Furthermore, PrPC has been shown to heighten spatial memory defects (13) and dendritic spine loss in AD mouse models (16). Furthermore, hippocampal primary culture and intrahippocampal injection indicate that the cytotoxic effects of Aβ oligomers are significantly reduced for PrP-null mice (17, 18). Similar results are seen in cell culture where cell lines lacking PrPC are resistant to Aβ toxicity (19–21). Aβ oligomers also influence PrPC trafficking and inhibit PrPC endocytosis (22). In addition, aggregates from AD brain extracts have been reported to contain Aβ bound to PrP (23, 24). Human genetic studies indicate that variants in PrP modify the risk for AD, specifically the Val129 polymorphism is protective, as compared with individuals homozygous for the Met129 allele (25). Such observations suggest that PrPC is an important acceptor/receptor for mediating the toxicity of Aβ (see review in Ref. 8).
The role of PrP in the Aβ-dependent pathogenesis of AD has proven controversial, not least because phenotypes may be generated in the absence of PrP in knock-out mice (26–29) and invertebrates that lack PrP (30, 31). It seems that not all Aβ toxicity is governed by PrPC; however, this does not preclude an important role for PrPC in mediating aspects of Aβ toxicity. Different Aβ assemblies and locations may well have different modes of toxic action; therefore these conflicting observations may simply reflect differences in genetic backgrounds and the nature of the Aβ-oligomers.
In mammals, PrPC is expressed at high levels and is concentrated at the synaptic terminals, anchored to the membrane by a glycosylphosphatidylinositol (GPI) moiety (32). Like Aβ, PrPC can also form amyloid fibers, and this conformational transition is linked to a range of transmissible spongiform encephalopathies in humans and mammals (33). Mammalian cellular prion proteins have a high structural and sequence homology and consist of two structurally distinct domains (34). The C-terminal domain (residues 126–231) is predominantly α-helical, whereas the Cu2+-binding N-terminal half of PrPC (residues 23–126) is natively disordered (35, 36).
The mechanism by which PrPC mediates Aβ toxicity is not well understood. In vitro, it is known that substoichiometric quantities of PrPC may trap Aβ as an oligomeric conformer and inhibit its progression to amyloid fibers (14). Furthermore, PrPC can also disassemble preformed Aβ40 fibrils (14) or favor lateral association of fibers (37). It appears that the unstructured N-terminal residues 23–126 are sufficient to inhibit Aβ fiber formation (14, 37, 38), although surprisingly others have indicated that the structured C terminus of PrPC inhibits fiber elongation (39). The binding of Aβ oligomers to PrPC at the cell surface may itself be sufficient to mediate Aβ toxicity; alternatively PrPC may also mediate Aβ's interactions with other proteins such as the N-methyl-d-aspartate receptor (40), Fyn kinase (16), and mGlu5 (41).
Our approach has been to investigate the interaction between Aβ and the mammalian PrPC by reconstituting a mammalian system in an organism that is normally naïve to both components. Extensive work (42–47) has shown that the quantitative measurement of Aβ-related phenotypes in flies, such as longevity and locomotor performance, correlates with the tendency of Aβ peptides to form oligomeric species (31, 48, 49). The fly model of Aβ toxicity has also provided insights into the mechanism of the sleep–wake dysrhythmia experienced by patients with AD (50). Similarly, the fly can be used to recreate PrP-mediated pathology (51), and furthermore, flies expressing ovine PrP are sensitive to scrapie brain extracts, and the PrPSc generated in the fly can transmit prion infection to other transgenic flies (52–54).
Combining both Aβ and PrP expression in the normally naïve brain of a fruit fly offers a powerful tool for measuring the potential of these proteins to show synergistic phenotypic enhancements. In particular, we show co-expression of Aβ and PrPC enhancing both longevity and circadian behavioral deficits in our fly model, under conditions where each protein alone has no effect.
Results
Co-expression of PrPC and Aβ results in reduced longevity in Drosophila
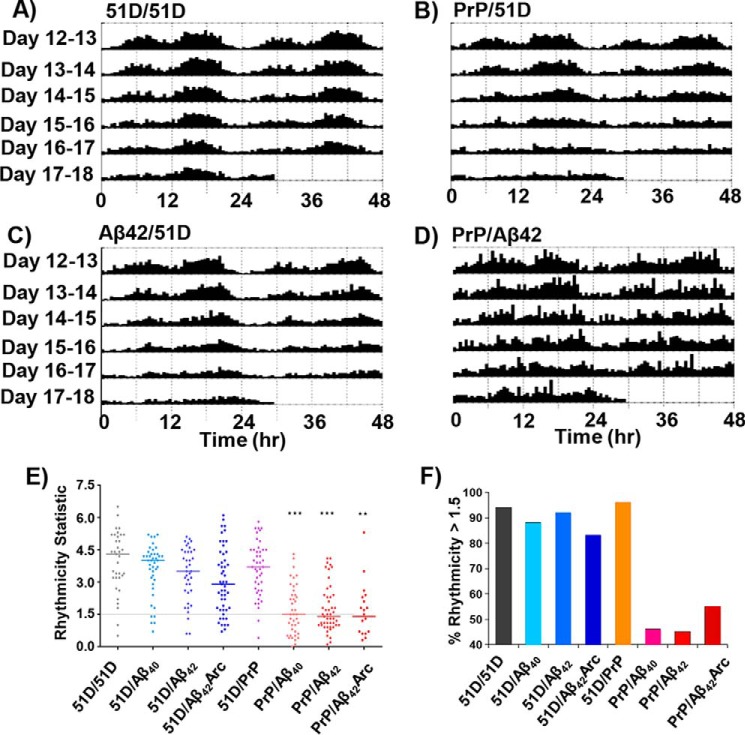
We used Drosophila as a model organism to look for in vivo interactions between Aβ and mammalian PrPC in a system that is normally naïve to both proteins. When flies have low levels of transgene expression—that is when cultured at 25 °C with single copy transgenes—neither the expression of mammalian PrPC nor the less neurotoxic Aβ isoforms (Aβ40 and Aβ42) alone reduced the lifespan of the flies. As expected, the highly aggregation prone Aβ42Arc (E22G arctic mutant) did reduce median survival by a third (Fig. 1 and Table 1). However, when PrPC was co-expressed with the same Aβ isoforms, in each case there was a significant decrease in the longevity of the flies. This effect was clearly synergistic for the combinations of PrPC with Aβ40 or Aβ42 (Fig. 1, A and B). In all cases the PrP-A-β interactions significantly reduced median survival as compared with the flies expressing either of the proteins alone, the effect being most marked for the interaction of PrPC with Aβ42 and Aβ42Arc (Fig. 1, B and C) where median survival times were halved (Table 1). The median survival times are presented in Fig. 1D where median data points represent vials of 10 flies, with a total of 400 flies/condition.
Figure 1.
Aβ and PrPC interact to reduce longevity in Drosophila. A–C, the effects on longevity when co-expressing PrPC with Aβ isoforms: Aβ40 (A), Aβ42 (B), and Aβ42Arc (C). The co-expression of PrP with Aβ (red), PrP only (orange), Aβ only (blue), and nontransgenic control flies 51D/51D (black) is shown. Co-expression of Aβ with PrP causes a significant reduction in longevity particularly for Aβ42 and Aβ42Arc flies. Each survival assay used 100 flies repeated four times; in total 400 flies/genotype were used. D, scatter plot showing the median survival of each vial of 10 flies, with 40 vials for each genotype. *, p < 0.05; ns, no significant difference.
Table 1.
Longevity assays for eight genotypes
The number of median days of survival is shown. 400 flies were used for each genotype. The standard deviation was calculated from variation in values for vials containing 10 flies (n = 40).
| 51D | PrPC | |
|---|---|---|
| 51D | 62 ± 7 | 65 ± 8 |
| Aβ40 | 70 ± 6 | 51 ± 8 |
| Aβ42 | 69 ± 5 | 36 ± 5 |
| Aβ42Arc | 44 ± 5 | 20 ± 2 |
The co-expression of Aβ with PrP disrupts circadian rhythm
One of the earliest features of AD and the fly model of Aβ toxicity is a progressive disruption in circadian rhythms, in particular the normal daily sleep–wake pattern. Previous studies have shown that high pan-neuronal Aβ expression in the fly results in sleep fragmentation, nocturnal locomotor activity, and a consequent reduction in the robustness of the circadian locomotor oscillation (50, 55). To look for an interaction between PrP and Aβ in such circadian phenotypes, we entrained flies for 3 days to a 12-h light:12-h dark cycle and then followed their circadian locomotor activity for a further 6 days in constant darkness using a beam-breaking actimeter. The expression of PrPC, Aβ40, and Aβ42 alone did not reduce the circadian locomotor cycle, with essentially all the flies retaining a robust rhythm (representative actograms for the interaction of PrPC and Aβ42 are shown in Fig. 2, A–D). The co-expression of PrPC with all Aβ isoforms resulted in a more marked reduction in rhythmicity in all cases (Fig. 2, E and F). In particular, the combination of PrPC with Aβ42 caused disrupted sleep–wake patterns in 55% of the flies. The highly aggregation prone Aβ42Arc, even in the absence of PrP, disrupted the cycle in 17% of flies.
Figure 2.
Aβ and PrPC interact to degrade circadian rhythmicity in a Drosophila model. A–D, four actograms showing the average locomotor activity of a group of typically 16 Drosophila males for each genotype. A, WT (51D/51D). B, PrP only. C, Aβ42 only. D, PrP + Aβ42. Flies were entrained in a 12-h light:12-h dark cycle up to day 12 and thereafter maintained in continuous darkness. The data are presented for days 12–18. E, rhythmicity statistic for the indicated genotypes and n number: 51D/51D, n = 35; 51D/Aβ40, n = 42; 51D/Aβ42, n = 38; 51D/Aβ42Arc, n = 51; 51D/PrP, n = 44; PrP/Aβ40, n = 41; PrP/Aβ42, n = 49; and PrP/Aβ42Arc, n = 20. Loss of rhythmicity is significant for the PrP + Aβ crossed flies. ***, p < 0.001; **, p < 0.01. F, the percentage of flies/genotype that retained circadian rhythmicity. The Aβ + PrP crossed flies (red shades) have marked loss in rhythmicity. The y axis shows the percentage of rhythmicity >1.5.
The presence of PrPC increases and localizes Aβ deposits
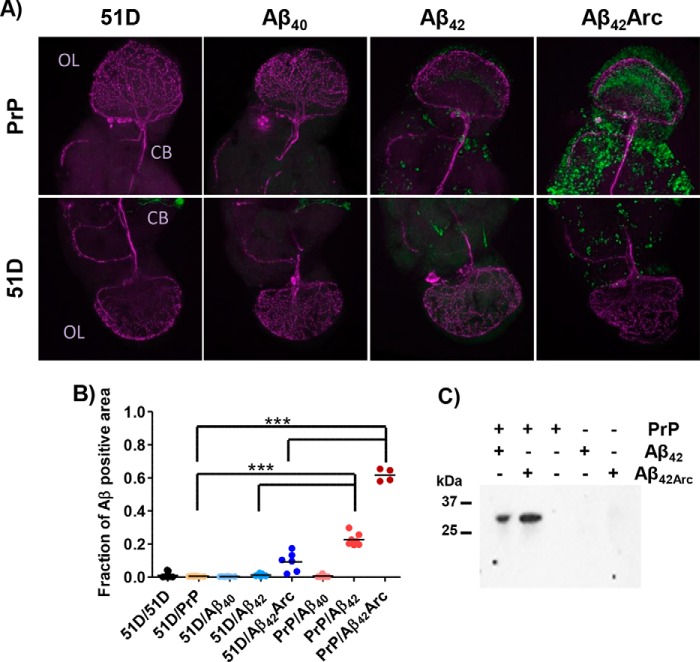
Next, we investigated how the expression of PrPC affects the build-up of Aβ deposits in Drosophila brain. We immunostained the brains of 20-day-old flies for Aβ deposits, looking for differences linked to the co-expression with PrPC. The 6E10 antibody (56) was used because its N-terminal epitope should be available in all Aβ conformers, including monomer, oligomer, and fibril. In the absence of PrPC, only Aβ42Arc showed some Aβ deposits, accounting for 10% of the area of the brain section. By contrast, the co-expression of PrPC boosted the deposition of Aβ42Arc greatly, resulting in positive staining over 60% of the section area. There was a synergistic interaction between PrPC and Aβ42, whereby neither of the transgenes alone resulted in any Aβ deposition, but together moderate levels of Aβ deposition occurred, accounting for >20% of the section area. The Aβ deposits are observed throughout the fly brain (Fig. 3).
Figure 3.
PrPC promotes Aβ accumulation in Drosophila brain by direct interaction. A, a comparison of Aβ-expressing flies with PrP (top row) or without PrP co-expression (bottom row). Anti-Aβ antibody (green) was used to identify all Aβ deposits, and an anti-pigment dispersing factor antibody (magenta) stains throughout fly brain including central brain (CB) and its optic lobe (OL); each image box is 250 × 250 μm. B, a plot presenting the fraction of Aβ positive area for Drosophila brain sections, 10 flies/construct. C, co-immunoprecipitation indicates direct Aβ–PrP interaction in the fly brain extracts. SDS Western blotting was carried out using SE16, the anti-Aβ antibody; the complex was precipitated using Sha31, the anti-PrP antibody. The Western blotting shown is for Aβ42–PrP and Aβ42Arc–PrP co-expressing flies, together with control flies expressing single transgenes.
The direct interaction of PrPC and Aβ in the fly brain was demonstrated by immunoprecipitation using the Sha31 anti-PrP antibody (57) with subsequent Western blotting with the 6E10 anti-Aβ antibody. Head extracts from flies expressing Aβ42 or Aβ42Arc, in the presence of PrPC, revealed a specific Aβ band on the Western blotting (Fig. 3C). Control experiments where either PrPC or Aβ was not expressed in the flies did not yield a band. The Aβ band suggests PrP binds to small SDS resistant oligomers.
In vitro, micromolar concentrations of PrP(23–231) inhibit Aβ fiber formation and fragment existing fibrils, promoting Aβ oligomers
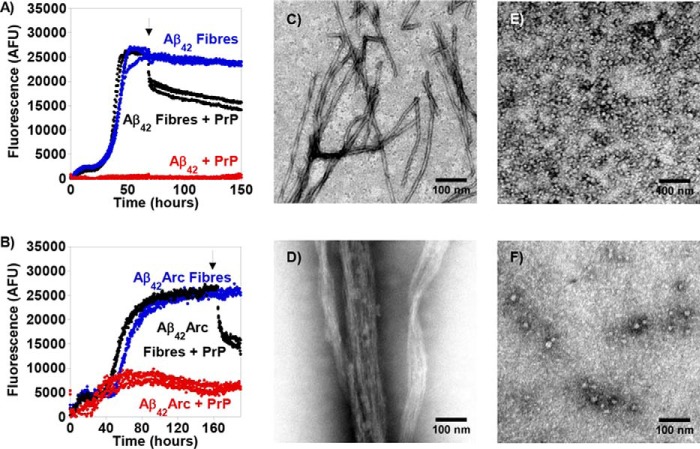
The amyloid-specific fluorescent dye thioflavin T (ThT) and transmission EM (TEM) were employed to probe the influence of PrPC on Aβ assembly in vitro. For 5 μm monomeric Aβ42 or Aβ42Arc, the time-dependent ThT signal was characteristic of a nucleated polymerization reaction consisting of an initial lag-phase followed by a rapid burst phase caused by amyloid fiber elongation (Fig. 4, A and B). At equilibrium, TEM images confirmed the presence of fibers for both Aβ42 (Fig. 4C) and Aβ42Arc (Fig. 4D). When 2.5 μm of full-length PrP(23–231) was added at the start of the reaction, the ThT signal was completely suppressed for Aβ42 and greatly reduced for Aβ42Arc (Fig. 4, A and B, red traces). Under these conditions, TEM revealed largely the presence of spherical oligomeric Aβ assemblies typically 10–20 nm in diameter (Fig. 4, E and F). Furthermore, the addition of 5 μm PrP(23–231) to a preparation of Aβ-amyloid fibers resulted in a 40–50% reduction in the ThT fluorescence signal, suggesting disruption of the preformed fibrils. TEM reveals a heterogeneous mixture of assembles; there were few mature fibrils. However, some structures appeared to be disrupted fibers, 10–20 nm wide and typically 100–200 nm long; other structures resembled circular Aβ oligomers similar to those in Fig. 4 (E and F). The fragmented short fiber rods tended to become laterally associated, a feature reported previously for short fragmented fibers (58, 59).
Figure 4.
In vitro Aβ42 and Aβ42Arc fiber formation inhibited by the presence of PrPC. A and B, fiber formation kinetics of Aβ42 and Aβ42Arc monitored by ThT fluorescence; in the absence of PrPC (blue) or in the presence of 0.5 molar equivalents of PrPC (red) and 1 molar equivalents of PrPC added to mature fibers (black). The arrow highlights the point at which PrPC was added to mature Aβ fibers. C and D, negative stain TEM images of Aβ42 fibers (C) and Aβ42Arc fibers (D). E and F, Aβ42 incubated in the presence of 0.5 molar equivalent of PrP(23–231) (E) and Aβ42Arc with PrPC (F). Scale bar, 100 nm.
Discussion
Although there is strong evidence from a number of laboratories to link some aspects of Aβ neurotoxicity and subsequent AD pathology with the presence of the cellular prion protein (PrPC) (8–10, 12, 13, 16, 40, 41), its precise role in AD pathology remains controversial (26, 29). There is a longstanding lack of consensus regarding the pathogenic role of PrPC within murine model systems; we therefore chose to use Drosophila as an alternative experimental organism. The Aβ-expressing fly has been shown to directly report oligomeric toxicity, principally through experiments correlating various quantitative phenotypes with the in vitro aggregation behavior of a range of Aβ isoforms (42–46). Likewise, the PrP-expressing flies have been well characterized by Thackray et al. (51), who have systematically investigated the cellular localizations of transgenic PrP in Drosophila. Notably, using immunocytochemistry, they show that WT mouse, hamster, and sheep PrP are all expressed on the surface of Drosophila S2 cells in culture. Using the same flies that we studied here, immunohistological examination of the PrP-expressing fly brain following scrapie prion infection is consistent with normal GPI-anchored surface expression and results in deposition of protease-resistant PrPSc along with vacuolation (53). They also show that altering the localization of the PrP, by removing the secretion signal peptide or the GPI anchor sequence, results in alterations in both neurotoxicity and prion susceptibility in this fly model (54). Thus, the fly has several advantages for studying the interaction of PrPC and Aβ, not least because there is no endogenous production of similar proteins. For this reason, the fly is unlikely to possess a conserved signaling pathway downstream of PrPC, potentially making the mechanism of any synergistic toxic interaction less complex. Specifically, the fly is well suited to the in vivo study of changes in the biophysical state of Aβ consequent upon its interaction with PrPC, which is anchored by GPI to the cellular membranes within in the brain of the fly (52).
The consistent finding in both the longevity and circadian behavioral assays is that the interactions between PrPC and both Aβ40 and Aβ42 are synergistic, because phenotypes only emerge when both proteins are co-expressed. Specifically, when the transgenes are expressed at low levels, lines expressing each protein alone show no deficits. Under the same conditions with both Aβ and PrPc expressed together, there is a marked reduction in longevity and loss of circadian rhythmicity. Importantly, the extent of synergistic interaction between Aβ and PrPC correlates well with the propensity of the various Aβ isoforms to spontaneously generate oligomeric assemblies in AD. Specifically, Aβ42 and Aβ42Arc peptides, by producing oligomeric ligands, are best suited to interact with PrPC, which results in a heightened phenotype.
Previous studies have established that Aβ oligomers bind PrPC with nanomolar affinity (9, 11). Here our co-immunoprecipitation studies show that Aβ and PrPC are interacting directly in the Drosophila brain. Aβ binds to the unstructured N terminus of PrPC (9, 11, 14, 37, 38), and substoichiometric PrPC will inhibit Aβ40 and Aβ42 fiber formation while promoting oligomer generation (14). We have previously shown that these PrPC trapped oligomers have a high β-sheet content and bind the Aβ-oligomer specific A11 antibody (14). These Aβ42 oligomers have been further characterized by cryo-TEM, which shows they form nanotubes (60). These cylindrical structures may be capable or penetrating the lipid bilayer to form toxic ion-channel pores (6). We show here PrPC will also inhibit the formation of fibrillar Aβ42Arc, favoring instead oligomeric assemblies, like those observed for Aβ42. There is a body of evidence indicating that small soluble Aβ42 oligomers rather than fibers are the cytotoxic form of Aβ (2–5), perhaps capable of forming ion channels (6). The ability of PrPC to act as a cell surface receptor for Aβ oligomers capable of trapping Aβ assemblies in an oligomeric form, in close proximity to the membrane, suggests a mechanism by which GPI membrane-anchored PrPC can help facilitate Aβ membrane interaction, which could then cause a loss of membrane integrity.
Remarkably, PrPC is capable of rapidly impacting the structure of preformed Aβ42 fibers, causing them to fragment. This fragmentation of fibers increases the number of fiber ends/unit length of fiber, which may increase cytotoxicity and is also likely to promote seeding of Aβ assemblies in the fly brain (61). The 6E10 antibody used to detect Aβ in the fly brain reports both oligomeric and fibrillar conformers; however, the increased deposition, with accompanying toxicity, is consistent with PrP-trapped oligomer formation. Such stabilization of oligomeric assemblies is supported by our in vitro experiments showing that for both Aβ42 and Aβ42Arc, the interaction with PrPC results in the generation of potentially toxic oligomeric aggregates irrespective of whether the initial Aβ conformer is monomeric or fibrillar. Taken together, this study predicts that PrPC potentiates AD pathogenesis. In the early stages, it encourages initial neurotoxic oligomer formation. Later in the disease, we would predict that the PrP–Aβ interaction should destabilize otherwise inert amyloid plaques, potentially releasing toxic amyloid fragments and favoring proteopathic seeding.
There is also a possibility that the presence of Aβ induces PrPC to misfold, self-associate, and become cytotoxic, although this is not seen in clinical specimens (62). Indeed, recently it has been shown that Aβ will instead inhibit PrP fiber formation (63). Furthermore, we show here that the degree of phenotype enhancement in the fly is determined by which Aβ isoform is present Aβ40 < Aβ42 < Aβ42Arc, indicating that the Aβ isoform influences toxicity in the fly model. This does preclude PrPSc driving the toxic action, but it is clear the co-expression of PrPC promotes Aβ deposition in the fly brain and Aβ oligomer formation in vitro.
A number of other amyloidogenic proteins can co-aggregate or influence assembly pathways of each other (64, 65). In particular both α-synuclein and islet amyloid polypeptide have been shown to interact with Aβ (66–68), while different isoforms of Aβ can also profoundly impact each other's fiber assembly (58, 59).
The loss of circadian rhythms and reduced life span of Aβ + PrP co-expressing flies strongly supports a role for PrPC in Aβ neurotoxicity in this model of AD. These data from Drosophila add to the murine studies and further support a crucial role for PrPC in the pathogenesis of AD. In vitro, PrPC binds to Aβ oligomers trapping Aβ assemblies in an oligomeric state, while Aβ fibers become fragmented. In vivo, this results in the accumulation of Aβ aggregates in the fly brain. Blocking the interaction between PrPC and Aβ may provide a novel avenue for AD therapy (60, 69–71).
Materials and methods
Drosophila lines
All Drosophila UAS-responsive transgenes were introduced into a w1118 background using pUAST/PhiC31-mediated site-directed transgenesis at the 51D acceptor site on the second chromosome (49). Flies expressing different Aβ peptides were generated as described by Crowther et al. (45). Three different constructs of Aβ isoform were used: Aβ40 (residues 1–40), Aβ42 (residues 1–42), and the Aβ42 arctic mutant (E22G) Aβ42Arc, associated with early onset familial AD. The elav c155-GAL4 system was employed to express human Aβ and the GPI-anchored ovine PrPC(23–231). The prion line was UAS-PrP, Val136–Arg154–Gln171 (VRQ) polymorphism as described by Thackray et al. (52). Flies expressing the transgenic proteins pan-neuronally were generated by crossing virgin female elavc155-GAL4 flies with male UAS-Aβ and UAS-PrP. These Aβ constructs expressed as the isolated peptide were co-expressed with the mammalian prion protein (PrPC) containing the VRQ isoform of ovine PrP.
We measured the mRNA levels for the single and co-expressed transgenes to confirm there was no down-regulation of Aβ or PrP observed. The β-actin levels were used as a standard for the quantitative RT-PCR experiments. For the quantitative RT-PCR and Western blotting studies, a total of 50 and 35 flies were used, respectively.
Fly rearing
All fly stocks and crossings were cultured in standard yeast cornmeal food vials. For each crossing five male and females were reared in the vial, with at 25 °C and 80% humidity with a 12-h light–dark (LD) cycle as described previously (45). Each day, newly eclosed offspring carrying a transgene for Aβ40, Aβ42, the E22G variant of Aβ42 (Aβ42Arc), and/or PrP (VRQ), driven by elavc155-GAL4 were placed in a fresh culture tube and incubated for a further 24 h at 25 °C to ensure that the females were mated. Control flies with WT X chromosomes (as a control for elavc155-GAL4) or the empty 51D transgenesis acceptor site chromosome (as a control for the transgenes) were handled in the same way. Following mating, the females were separated and retained for longevity assays, whereas the males were used for circadian actimetry assays.
Longevity assays
Longevity assays were performed using 100 mated female flies as described previously (45). Each assay was repeated four times using flies from independent genetic crosses; a total of 400 flies were used in each comparison. Surviving flies were counted and placed in fresh culture tubes twice per week. The data were visualized using Kaplan–Meier survival plots, and the statistical significance of any differences in longevity was estimate using the Student's t test, calculated using n = 40 estimations of the population median survival. A p value of <0.05 was set as the threshold for statistical significance. Differences in longevity were analyzed using the log-rank statistical method, using Prism software.
Circadian locomotor
Adult male flies of each genotype were aged to 9 days after eclosion before being transferred to individual tubes and entrained for 3 days using 12-h light:12-h dark cycles. For the subsequent 6 days, the fly locomotor activity was monitored in constant darkness by counting the breaks of an IR beam within the fly tube, with counts summed every 30 min (DAM monitoring system; Trikinetics, Waltham, MA). The Drosophila circadian assay has been described previously (50, 74). The rhythmic percentage is the fraction of flies that achieve a rhythmicity statistic >1.5. We quantify the strength of the circadian rhythm using the rhythmicity index. The rhythmicity index is obtained from the height of the third peak on the correlogram (75, 76). The ratio of the rhythmicity index value compared with the 95% confidence line gives the rhythmicity statistic value (77). The average actograms for 16 flies were plotted using the Flytoolbox on MATLAB for all light–dark and constant-darkness sessions.
Immunohistochemistry
Protein accumulation in fly brains were imaged by fluorescence microscopy using the anti-Aβ1–16 (6E10), anti-PDF (pigment dispersing factor), and Anti-PrP (Sha31) antibodies. For each condition, 10 female flies were fixed in 4% w/v paraformaldehyde (0.1 m phosphate buffer, pH 7.4, with 0.1% v/v Triton X-100) at an age of 20 days after eclosion. After 3 h of fixation at room temperature, the flies were washed three times with phosphate buffer at room temperature. The Drosophila brains were then dissected on ice, and nonspecific protein binding was blocked with 10% (w/v) goat serum (in phosphate buffer with 0.5% v/v Triton X-100) for 2 h at room temperature. The brains were then stained using two primary antibodies: monoclonal mouse anti-PDF (1:1000, PDFC7; Developmental Studies Hybridoma Bank) and polyclonal rabbit anti-Aβ1–16 (1:1000, SIG-39322; Covance) at 4 °C for 24 h. Six steps of washing (in phosphate buffer with 0.5% v/v Triton X-100) followed by incubation with Alexa Fluor 488–conjugated anti-mouse and Alexa Fluor 647–conjugated anti-rabbit antibodies (both at 1:500) overnight at 4 °C. Finally, the brains were washed six more times, mounted in Vectashield (Vector Laboratories), and stored at 4 °C until viewed under a Nikon Eclipse C1si confocal microscope. This protocol is modified from that described by Hermann et al. (78). The percentage stained parameter was calculated using ImageJ and reports the fraction of Drosophila brain section area that stain for Aβ.
Co-immunoprecipitation of Aβ with PrP
Approximately a thousand flies or more (3 cm3), expressing Aβ42 or Aβ42Arc with and without PrP, were flash frozen in liquid nitrogen and decapitated by vortexing while frozen. For each genotype, 400 μl of isolated Drosophila heads were homogenized in 400 μl of extraction buffer (20 mm HEPES, pH 7.5, 100 mm KCl, 1 mm DTT, 5% (v/v) glycerol, 0.05% (v/v) Nonidet P40, Roche cOmplete protease inhibitor) using a snap-top tube and disposable pestel. This fly homogenate was precleared by centrifugation at 2,000 × g for 20 min to remove Aβ and PrP aggregates. Separately 20 μl of protein G–Sepharose fast flow beads (Amersham Biosciences) were first incubated with anti-PrP antibody (Sha31, 5 μl of undiluted). The beads were incubated with the antibody for 1 h at 4 °C, with gentle agitation. The beads were then washed twice with incubation buffer and added to the Drosophila head extracts for 16 h at 4 °C. The beads were washed twice with extraction buffer before being resuspended in 5 μl of lithium dodecyl sulfate loading buffer. The proteins were separated on a 4–20% (w/v) gradient SDS–PAGE gel and blotted onto a nitrocellulose membrane. The membrane was blocked in PBS containing 5% (w/v) nonfat milk for 1 h at room temperature and incubated with horseradish peroxidase-conjugated rabbit polyclonal anti-Aβ1–16 (1:1000, SIG-39322; Covance) for 1 h at room temperature. After six washes in PBS, 0.05% (v/v) Triton, the blot was incubated with a goat anti-rabbit secondary antibody and incubated at room temperature for 90 min. The washing steps were repeated, and the membrane was developed using the SuperSignal West Femto kit. Transgene expression levels were similar across the genotypes, as determined by qPCR. However, the Aβ interaction with PrP results in the accumulation of Aβ; for this reason, input levels of Aβ were not measured. The loading of the Western blotting was based on equal tissue extract per lane.
Recombinant Syrian hamster PrP expression and purification
Tag-free, full-length Syrian hamster PrP, SHaPrP-(23–231), was cloned into a pET-23 vector and expressed in Rosetta (DE3)pLysS Escherichia coli cells. Cultures were grown at 37 °C to absorption at 600 nm of 0.9 AU, and protein expression was induced by 1 mm isopropyl β-d-1-thiogalactopyranoside. The cells were grown for a further 16–18 h at 16 °C. Bacterial pellets were lysed by sonication in resuspension buffer (150 mm NaCl and 50 mm Tris-HCl, pH 7.5). The soluble fraction was removed by centrifugation (20,000 × g for 15 min), and the insoluble fraction was resuspended in solubilization buffer (8 m urea, 150 mm NaCl, and 50 mm Tris HCl, pH 7.5). Cell debris was removed by centrifugation (20,000 × g for 15 min), and the soluble fraction was applied to chelating Sepharose (Amersham Biosciences) charged with copper. The Sepharose was washed with five column volumes of solubilization buffer and refolded in wash buffer (100 mm NaCl, 10 mm imidazole, 50 mm Tris-HCl, pH 7.5) containing decreasing concentrations of urea. The protein was eluted from the column (100 mm NaCl, 0.5 m imidazole, 50 mm Tris-HCl, pH 7.5) and concentrated in a centrifugal concentrator (Sartorius) with a 10-kDa molecular mass cutoff. The protein was further purified by size exclusion chromatography using an S75 column (10/300 GL) attached to an AKTA PURE (GE Healthcare). The protein was assessed to be greater than 95% pure by SDS-PAGE (4–20% Tris-glycine gel; Bio-Rad) and stained with InstantBlue (Expedeon). The concentration of ShaPrP(23–231) was measured by its absorbance at 280 nm using a molar absorption coefficient of 62,280 m−1 cm−1.
Aβ peptide
Aβ peptides were purchased from Cambridge Research Biochemicals or EZBiolab, synthesized using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry, purified as a single peak on HPLC and characterized by MS. The purchased peptides included human β-amyloid peptide, residues 1–42, and the arctic mutant E22G; designated Aβ42 and Aβ42Arc, respectively. Aβ42 and Aβ42Arc were solubilized at 0.7 mg/ml in water at pH 10 with gentle rocking at 4 °C for 4 h, maintaining pH 10 by addition of NaOH. Aβ42 and Aβ42Arc were purified using size-exclusion chromatography after solubilization using an S200 column, a protocol adapted from (72). The absorbance at 280 nm was used to calculate the concentration of Aβ, with an extinction coefficient of 1280 m−1 cm−1.
Fiber growth kinetics
Fiber growth kinetics was monitored using a 96-well plate with ThT, a fiber-specific fluorescent dye. Solubilized Aβ42 and Aβ42Arc was incubated at 30 °C with 30 mm HEPES, pH 7.4, and 160 mm NaCl, with intermittent shaking in the presence and absence of recombinant PrPC(23–231). The growth of Aβ fibers was monitored using a 96-well microplate plate and a BMG Omega FLUOstar fluorescence reader, with an excitation filter at 440 nm and an emission filter at 490 nm. Each reading consists of 20 flashes, prior to each reading, the plate was agitated for 30 s with double orbital (200 rpm) shaking every 30 min. Sterile flat-bottomed plates were used and sealed with Starseal polyolefin sealing film. A volume of 200 μl was used per well. The pH of a sample was monitored before and after each experiment; a variation of ±0.05 pH units or less was observed over the course of the experiment. ThT additions were made from a fresh 2 mm stock solution in water. The fibril growth experiments were typically carried out using 5 μm monomeric Aβ42 and Aβ42Arc in the presence of 2 molar equivalents of ThT (73). The sample was incubated at 30 °C in the presence of 160 mm NaCl, 30 mm HEPES, pH 7.4. Fluorescence measurements were made from above, using an orbital averaging sample reading (4-mm diameter). For the disaggregation experiments, small aliquots of ShaPrP(23–231) were added to preformed fibrillary Aβ, whereas the 96-well plate was left on the plate holder within the reader.
Transmission electron microscopy (TEM)
A negative stain of phosphotungstinic acid was used to image Aβ fiber/oligomers assemblies using TEM. Carbon-coated 300-mesh copper grids (SPI Supplies) were glow discharged at the start of each experiment. 5-μl aliquots of the 5 μm Aβ sample were absorbed onto each grid for 90 s before blotting dry. The grids were washed with water followed by incubation with 5 μl of 2% (w/v) phosphotungstic acid, pH 7.4, for 70 s, to produce a negatively stained Aβ. Finally the grids were washed in water. Images of the grids were recorded on a JEOL JEM 1230 electron microscope operated at 80 kV, a Morada 2k CCD camera system, and the iTEM software package (Olympus Europa).
Author contributions
N. D. Y., K.-F. C., D. C. C., and J. H. V. formal analysis; N. D. Y., R.-S. R., and D. C. C. investigation; N. D. Y. visualization; N. D. Y., R.-S. R., and D. C. C. methodology; N. D. Y. writing-original draft; N. D. Y., K.-F. C., R.-S. R., D. C. C., and J. H. V. writing-review and editing; D. C. C. and J. H. V. conceptualization; D. C. C. data curation; D. C. C. and J. H. V. supervision; D. C. C. and J. H. V. funding acquisition; D. C. C. and J. H. V. project administration.
Acknowledgments
Syrian hamster PrP pET-23 vector was a gift from T. Pinhero (Warwick University). We thank Drs. Bujdoso and Thackray (University of Cambridge) for the UAS-PrP Drosophila lines.
This work was supported by Wellcome Trust Grant 093241/Z/10/Z and Biotechnology and Biological Sciences Research Council Grant BB/M023877/1. The authors declare that they have no conflicts of interest with the contents of this article.
- AD
- Alzheimer's disease
- Aβ
- amyloid β peptide
- Aβ40
- Aβ residues 1–40
- Aβ42
- Aβ residues 1–42
- Aβ42Arc
- Aβ42 arctic mutant (E22G)
- PrP
- prion protein
- PrPC(23–231)
- cellular prion protein residues 23–231
- TEM
- transmission electron microscopy
- ThT
- thioflavin
- GPI
- glycosylphosphatidylinositol.
References
- 1. Prince M., Bryce R., Albanese E., Wimo A., Ribeiro W., and Ferri C. P. (2013) The global prevalence of dementia: a systematic review and metaanalysis. Alzheimers Dementia 9, 63–75 10.1016/j.jalz.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 2. Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., and Selkoe D. J. (2002) Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416, 535–539 10.1038/416535a [DOI] [PubMed] [Google Scholar]
- 3. Lambert M. P., Barlow A. K., Chromy B. A., Edwards C., Freed R., Liosatos M., Morgan T. E., Rozovsky I., Trommer B., Viola K. L., Wals P., Zhang C., Finch C. E., Krafft G. A., and Klein W. L. (1998) Diffusible, nonfibrillar ligands derived from Aβ1–42 are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. U.S.A. 95, 6448–6453 10.1073/pnas.95.11.6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lesné S., Koh M. T., Kotilinek L., Kayed R., Glabe C. G., Yang A., Gallagher M., and Ashe K. H. (2006) A specific amyloid-β protein assembly in the brain impairs memory. Nature 440, 352–357 10.1038/nature04533 [DOI] [PubMed] [Google Scholar]
- 5. Yankner B. A., and Lu T. (2009) Amyloid β-protein toxicity and the pathogenesis of Alzheimer disease. J. Biol. Chem. 284, 4755–4759 10.1074/jbc.R800018200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bode D. C., Baker M. D., and Viles J. H. (2016) Ion channel formation by amyloid-β42 oligomers but not amyloid-β40 in cellular membranes. J. Biol. Chem. 292, 1404–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jarosz-Griffiths H. H., Noble E., Rushworth J. V., and Hooper N. M. (2016) Amyloid-β receptors: the good, the bad, and the prion protein. J. Biol. Chem. 291, 3174–3183 10.1074/jbc.R115.702704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Purro S. A., Nicoll A. J., and Collinge J. (2018) Prion protein as a toxic acceptor of amyloid-β oligomers. Biol. Psychiatry 83, 358–368 10.1016/j.biopsych.2017.11.020 [DOI] [PubMed] [Google Scholar]
- 9. Laurén J., Gimbel D. A., Nygaard H. B., Gilbert J. W., and Strittmatter S. M. (2009) Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature 457, 1128–1132 10.1038/nature07761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barry A. E., Klyubin I., Mc Donald J. M., Mably A. J., Farrell M. A., Scott M., Walsh D. M., and Rowan M. J. (2011) Alzheimer's disease brain-derived amyloid-β–mediated inhibition of LTP in vivo is prevented by immunotargeting cellular prion protein. J. Neurosci. 31, 7259–7263 10.1523/JNEUROSCI.6500-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen S., Yadav S. P., and Surewicz W. K. (2010) Interaction between human prion protein and amyloid-β (Aβ) oligomers: role of N-terminal residues. J. Biol. Chem. 285, 26377–26383 10.1074/jbc.M110.145516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Freir D. B., Nicoll A. J., Klyubin I., Panico S., Mc Donald J. M., Risse E., Asante E. A., Farrow M. A., Sessions R. B., Saibil H. R., Clarke A. R., Rowan M. J., Walsh D. M., and Collinge J. (2011) Interaction between prion protein and toxic amyloid β assemblies can be therapeutically targeted at multiple sites. Nat. Commun. 2, 336 10.1038/ncomms1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gimbel D. A., Nygaard H. B., Coffey E. E., Gunther E. C., Lauren J., Gimbel Z. A., and Strittmatter S. M. (2010) Memory impairment in transgenic Alzheimer mice requires cellular prion protein. J. Neurosci. 30, 6367–6374 10.1523/JNEUROSCI.0395-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Younan N. D., Sarell C. J., Davies P., Brown D. R., and Viles J. H. (2013) The cellular prion protein traps Alzheimer's Aβ in an oligomeric form and disassembles amyloid fibers. FASEB J. 27, 1847–1858 10.1096/fj.12-222588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dohler F., Sepulveda-Falla D., Krasemann S., Altmeppen H., Schlüter H., Hildebrand D., Zerr I., Matschke J., and Glatzel M. (2014) High molecular mass assemblies of amyloid-β oligomers bind prion protein in patients with Alzheimer's disease. Brain 137, 873–886 10.1093/brain/awt375 [DOI] [PubMed] [Google Scholar]
- 16. Um J. W., Nygaard H. B., Heiss J. K., Kostylev M. A., Stagi M., Vortmeyer A., Wisniewski T., Gunther E. C., and Strittmatter S. M. (2012) Alzheimer amyloid-β oligomer bound to postsynaptic prion protein activates Fyn to impair neurons. Nat. Neurosci. 15, 1227–1235 10.1038/nn.3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bate C., and Williams A. (2011) Amyloid-β-induced synapse damage is mediated via cross-linkage of cellular prion proteins. J. Biol. Chem. 286, 37955–37963 10.1074/jbc.M111.248724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kudo W., Lee H. P., Zou W. Q., Wang X., Perry G., Zhu X., Smith M. A., Petersen R. B., and Lee H. G. (2012) Cellular prion protein is essential for oligomeric amyloid-β-induced neuronal cell death. Hum. Mol. Genet. 21, 1138–1144 10.1093/hmg/ddr542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Resenberger U. K., Harmeier A., Woerner A. C., Goodman J. L., Muller V., Krishnan R., Vabulas R. M., Kretzschmar H. A., Lindquist S., Hartl F. U., Multhaup G., Winklhofer K. F., and Tatzelt J. (2011) The cellular prion protein mediates neurotoxic signalling of β-sheet-rich conformers independent of prion replication. EMBO J. 30, 2057–2070 10.1038/emboj.2011.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Resenberger U. K., Winklhofer K. F., and Tatzelt J. (2012) Cellular prion protein mediates toxic signaling of amyloid β. Neurodegener Dis. 10, 298–300 10.1159/000332596 [DOI] [PubMed] [Google Scholar]
- 21. Pinnock E. C., Jovanovic K., Pinto M. G., Ferreira E., Dias Bda C., Penny C., Knackmuss S., Reusch U., Little M., Schatzl H. M., and Weiss S. F. (2016) LRP/LR antibody mediated rescuing of amyloid-β–induced cytotoxicity is dependent on PrPc in Alzheimer's disease. J. Alzheimer's Dis. 49, 645–657 [DOI] [PubMed] [Google Scholar]
- 22. Caetano F. A., Beraldo F. H., Hajj G. N., Guimaraes A. L., Jürgensen S., Wasilewska-Sampaio A. P., Hirata P. H., Souza I., Machado C. F., Wong D. Y., De Felice F. G., Ferreira S. T., Prado V. F., Rylett R. J., Martins V. R., et al. (2011) Amyloid-β oligomers increase the localization of prion protein at the cell surface. J. Neurochem. 117, 538–553 10.1111/j.1471-4159.2011.07225.x [DOI] [PubMed] [Google Scholar]
- 23. Zou W. Q., Xiao X., Yuan J., Puoti G., Fujioka H., Wang X., Richardson S., Zhou X., Zou R., Li S., Zhu X., McGeer P. L., McGeehan J., Kneale G., Rincon-Limas D. E., et al. (2011) Amyloid β interacts mainly with insoluble prion protein in the Alzheimer brain. J. Biol. Chem. 286, 15095–15105 10.1074/jbc.M110.199356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ferrer I., Blanco R., Carmona M., Puig B., Ribera R., Rey M. J., and Ribalta T. (2001) Prion protein expression in senile plaques in Alzheimer's disease. Acta Neuropathol. 101, 49–56 [DOI] [PubMed] [Google Scholar]
- 25. He J., Li X., Yang J., Huang J., Fu X., Zhang Y., and Fan H. (2013) The association between the methionine/valine (M/V) polymorphism (rs1799990) in the PRNP gene and the risk of Alzheimer disease: an update by meta-analysis. J. Neurol. Sci. 326, 89–95 10.1016/j.jns.2013.01.020 [DOI] [PubMed] [Google Scholar]
- 26. Balducci C., Beeg M., Stravalaci M., Bastone A., Sclip A., Biasini E., Tapella L., Colombo L., Manzoni C., Borsello T., Chiesa R., Gobbi M., Salmona M., and Forloni G. (2010) Synthetic amyloid-β oligomers impair long-term memory independently of cellular prion protein. Proc. Natl. Acad. Sci. U.S.A. 107, 2295–2300 10.1073/pnas.0911829107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Calella A. M., Farinelli M., Nuvolone M., Mirante O., Moos R., Falsig J., Mansuy I. M., and Aguzzi A. (2010) Prion protein and Aβ-related synaptic toxicity impairment. EMBO Mol. Med. 2, 306–314 10.1002/emmm.201000082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kessels H. W., Nguyen L. N., Nabavi S., and Malinow R. (2010) The prion protein as a receptor for amyloid-β. Nature 466, E3–E5 10.1038/nature09217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cissé M., Sanchez P. E., Kim D. H., Ho K., Yu G. Q., and Mucke L. (2011) Ablation of cellular prion protein does not ameliorate abnormal neural network activity or cognitive dysfunction in the J20 line of human amyloid precursor protein transgenic mice. J. Neurosci. 31, 10427–10431 10.1523/JNEUROSCI.1459-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Link C. D. (1995) Expression of human β-amyloid peptide in transgenic Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 92, 9368–9372 10.1073/pnas.92.20.9368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Luheshi L. M., Tartaglia G. G., Brorsson A. C., Pawar A. P., Watson I. E., Chiti F., Vendruscolo M., Lomas D. A., Dobson C. M., and Crowther D. C. (2007) Systematic in vivo analysis of the intrinsic determinants of amyloid β pathogenicity. PLoS Biol. 5, e290 10.1371/journal.pbio.0050290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Herms J., Tings T., Gall S., Madlung A., Giese A., Siebert H., Schürmann P., Windl O., Brose N., and Kretzschmar H. (1999) Evidence of presynaptic location and function of the prion protein. J. Neurosci. 19, 8866–8875 10.1523/JNEUROSCI.19-20-08866.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Prusiner S. B. (1998) Prions. Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 10.1073/pnas.95.23.13363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Donne D. G., Viles J. H., Groth D., Mehlhorn I., James T. L., Cohen F. E., Prusiner S. B., Wright P. E., and Dyson H. J. (1997) Structure of the recombinant full-length hamster prion protein PrP(29–231): the N terminus is highly flexible. Proc. Natl. Acad. Sci. U.S.A. 94, 13452–13457 10.1073/pnas.94.25.13452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Viles J. H., Donne D., Kroon G., Prusiner S. B., Cohen F. E., Dyson H. J., and Wright P. E. (2001) Local structural plasticity of the prion protein: analysis of NMR relaxation dynamics. Biochemistry 40, 2743–2753 10.1021/bi002898a [DOI] [PubMed] [Google Scholar]
- 36. O'Sullivan D. B., Jones C. E., Abdelraheim S. R., Brazier M. W., Toms H., Brown D. R., and Viles J. H. (2009) Dynamics of a truncated prion protein, PrP(113–231), from 15N NMR relaxation: order parameters calculated and slow conformational fluctuations localized to a distinct region. Protein Sci. 18, 410–423 10.1002/pro.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nieznanski K., Surewicz K., Chen S., Nieznanska H., and Surewicz W. K. (2014) Interaction between prion protein and Aβ amyloid fibrils revisited. ACS Chem. Neurosci. 5, 340–345 10.1021/cn500019c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nieznanski K., Choi J. K., Chen S., Surewicz K., and Surewicz W. K. (2012) Soluble prion protein inhibits amyloid-β (Aβ) fibrillization and toxicity. J. Biol. Chem. 287, 33104–33108 10.1074/jbc.C112.400614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bove-Fenderson E., Urano R., Straub J. E., and Harris D. A. (2017) Cellular prion protein targets amyloid-β fibril ends via its C-terminal domain to prevent elongation. J. Biol. Chem. 292, 16858–16871 10.1074/jbc.M117.789990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. You H., Tsutsui S., Hameed S., Kannanayakal T. J., Chen L., Xia P., Engbers J. D., Lipton S. A., Stys P. K., and Zamponi G. W. (2012) Aβ neurotoxicity depends on interactions between copper ions, prion protein, and N-methyl-d-aspartate receptors. Proc. Natl. Acad. Sci. U.S.A. 109, 1737–1742 10.1073/pnas.1110789109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu N.-W., Nicoll A. J., Zhang D., Mably A. J., O'Malley T., Purro S. A., Terry C., Collinge J., Walsh D. M., and Rowan M. J. (2014) mGlu5 receptors and cellular prion protein mediate amyloid-β-facilitated synaptic long-term depression in vivo. Nat. Commun. 5, 3374 10.1038/ncomms4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Iijima K., Liu H.-P., Chiang A.-S., Hearn S. A., Konsolaki M., and Zhong Y. (2004) Dissecting the pathological effects of human Aβ40 and Aβ42 in Drosophila: a potential model for Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 101, 6623–6628 10.1073/pnas.0400895101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Greeve I., Kretzschmar D., Tschäpe J.-A., Beyn A., Brellinger C., Schweizer M., Nitsch R. M., and Reifegerste R. (2004) Age-dependent neurodegeneration and Alzheimer-amyloid plaque formation in transgenic Drosophila. J. Neurosci. 24, 3899–3906 10.1523/JNEUROSCI.0283-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Finelli A., Kelkar A., Song H.-J., Yang H., and Konsolaki M. (2004) A model for studying Alzheimer's Aβ42-induced toxicity in Drosophila melanogaster. Mol. Cell. Neurosci. 26, 365–375 10.1016/j.mcn.2004.03.001 [DOI] [PubMed] [Google Scholar]
- 45. Crowther D. C., Kinghorn K. J., Miranda E., Page R., Curry J. A., Duthie F. A., Gubb D. C., and Lomas D. A. (2005) Intraneuronal Aβ, non-amyloid aggregates and neurodegeneration in a Drosophila model of Alzheimer's disease. Neuroscience 132, 123–135 10.1016/j.neuroscience.2004.12.025 [DOI] [PubMed] [Google Scholar]
- 46. Stokin G. B., Almenar-Queralt A., Gunawardena S., Rodrigues E. M., Falzone T., Kim J., Lillo C., Mount S. L., Roberts E. A., McGowan E., Williams D. S., and Goldstein L. S. (2008) Amyloid precursor protein-induced axonopathies are independent of amyloid-β peptides. Hum. Mol. Genet. 17, 3474–3486 10.1093/hmg/ddn240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moloney A., Sattelle D. B., Lomas D. A., and Crowther D. C. (2010) Alzheimer's disease: insights from Drosophila melanogaster models. Trends Biochem. Sci. 35, 228–235 10.1016/j.tibs.2009.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Iijima K., Chiang H. C., Hearn S. A., Hakker I., Gatt A., Shenton C., Granger L., Leung A., Iijima-Ando K., and Zhong Y. (2008) Aβ42 mutants with different aggregation profiles induce distinct pathologies in Drosophila. PLoS One 3, e1703 10.1371/journal.pone.0001703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Speretta E., Jahn T. R., Tartaglia G. G., Favrin G., Barros T. P., Imarisio S., Lomas D. A., Luheshi L. M., Crowther D. C., and Dobson C. M. (2012) Expression in drosophila of tandem amyloid β peptides provides insights into links between aggregation and neurotoxicity. J. Biol. Chem. 287, 20748–20754 10.1074/jbc.M112.350124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen K.-F., Possidente B., Lomas D. A., and Crowther D. C. (2014) The central molecular clock is robust in the face of behavioural arrhythmia in a Drosophila model of Alzheimer's disease. Dis. Model Mech. 7, 445–458 10.1242/dmm.014134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thackray A. M., Cardova A., Wolf H., Pradl L., Vorberg I., Jackson W. S., and Bujdoso R. (2017) Genetic human prion disease modelled in PrP transgenic Drosophila. Biochem. J. 474, 3253–3267 10.1042/BCJ20170462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thackray A. M., Muhammad F., Zhang C., Di Y., Jahn T. R., Landgraf M., Crowther D. C., Evers J. F., and Bujdoso R. (2012) Ovine PrP transgenic Drosophila show reduced locomotor activity and decreased survival. Biochem. J. 444, 487–495 10.1042/BJ20112141 [DOI] [PubMed] [Google Scholar]
- 53. Thackray A. M., Muhammad F., Zhang C., Denyer M., Spiropoulos J., Crowther D. C., and Bujdoso R. (2012) Prion-induced toxicity in PrP transgenic Drosophila. Exp. Mol. Pathol. 92, 194–201 10.1016/j.yexmp.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 54. Thackray A. M., Di Y., Zhang C., Wolf H., Pradl L., Vorberg I., Andréoletti O., and Bujdoso R. (2014) Prion-induced and spontaneous formation of transmissible toxicity in PrP transgenic Drosophila. Biochem. J. 463, 31–40 10.1042/BJ20140129 [DOI] [PubMed] [Google Scholar]
- 55. Long D. M., Blake M. R., Dutta S., Holbrook S. D., Kotwica-Rolinska J., Kretzschmar D., and Giebultowicz J. M. (2014) Relationships between the circadian system and Alzheimer's disease-like symptoms in Drosophila. PLoS One 9, e106068 10.1371/journal.pone.0106068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim K., Wen G., Bancher C., Chen C., Sapienza V., Hong H., and Wisniewski H. (1990) Detection and quantitation of amyloid b-peptide with 2 monoclonal-antibodies. Neurosci. Res. Commun. 7, 113–122 [Google Scholar]
- 57. Morel N., Simon S., Frobert Y., Volland H., Mourton-Gilles C., Negro A., Sorgato M. C., Créminon C., and Grassi J. (2004) Selective and efficient immunoprecipitation of the disease-associated form of the prion protein can be mediated by nonspecific interactions between monoclonal antibodies and scrapie-associated fibrils. J. Biol. Chem. 279, 30143–30149 10.1074/jbc.M403896200 [DOI] [PubMed] [Google Scholar]
- 58. Barritt J. D., Younan N. D., and Viles J. H. (2017) N-terminally truncated amyloid-β(11–40/42) cofibrillizes with its full-length counterpart: implications for Alzheimer's disease. Angew. Chem. Int. Ed. Engl. 56, 9816–9819 10.1002/anie.201704618 [DOI] [PubMed] [Google Scholar]
- 59. Barritt J. D., and Viles J. H. (2015) Truncated amyloid-β(11–40/42) from Alzheimer disease binds Cu2+ with a femtomolar affinity and influences fiber assembly. J. Biol. Chem. 290, 27791–27802 10.1074/jbc.M115.684084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nicoll A. J., Panico S., Freir D. B., Wright D., Terry C., Risse E., Herron C. E., O'Malley T., Wadsworth J. D., Farrow M. A., Walsh D. M., Saibil H. R., and Collinge J. (2013) Amyloid-β nanotubes are associated with prion protein-dependent synaptotoxicity. Nat. Commun. 4, 2416 10.1038/ncomms3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sowade R. F., and Jahn T. R. (2017) Seed-induced acceleration of amyloid-β mediated neurotoxicity in vivo. Nat. Commun. 8, 512 10.1038/s41467-017-00579-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rahimi J., and Kovacs G. G. (2014) Prevalence of mixed pathologies in the aging brain. Alzheimers Res. Ther. 6, 82 10.1186/s13195-014-0082-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sarell C. J., Quarterman E., Yip D. C., Terry C., Nicoll A. J., Wadsworth J. D. F., Farrow M. A., Walsh D. M., and Collinge J. (2017) Soluble A β aggregates can inhibit prion propagation. Open Biol. 7, 170158 10.1098/rsob.170158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Luo J., Wärmländer S. K., Gräslund A., and Abrahams J. P. (2016) Cross-interactions between the Alzheimer disease amyloid-β peptide and other amyloid proteins: a further aspect of the amyloid cascade hypothesis. J. Biol. Chem. 291, 16485–16493 10.1074/jbc.R116.714576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sarell C. J., Stockley P. G., and Radford S. E. (2013) Assessing the causes and consequences of co-polymerization in amyloid formation. Prion 7, 359–368 10.4161/pri.26415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bachhuber T., Katzmarski N., McCarter J. F., Loreth D., Tahirovic S., Kamp F., Abou-Ajram C., Nuscher B., Serrano-Pozo A., Müller A., Prinz M., Steiner H., Hyman B. T., Haass C., and Meyer-Luehmann M. (2015) Inhibition of amyloid-β plaque formation by alpha-synuclein. Nat. Med. 21, 802–807 10.1038/nm.3885 [DOI] [PubMed] [Google Scholar]
- 67. Clinton L. K., Blurton-Jones M., Myczek K., Trojanowski J. Q., and LaFerla F. M. (2010) Synergistic Interactions between Aβ, τ, and α-synuclein: acceleration of neuropathology and cognitive decline. J. Neurosci. 30, 7281–7289 10.1523/JNEUROSCI.0490-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yan L. M., Velkova A., Tatarek-Nossol M., Andreetto E., and Kapurniotu A. (2007) LAPP mimic blocks A β cytotoxic self-assembly: cross-suppression of amyloid toxicity of A β and IAPP suggests a molecular link between Alzheimer's disease and type II diabetes. Angew. Chem. Int. Edit. Engl. 46, 1246–1252 10.1002/anie.200604056 [DOI] [PubMed] [Google Scholar]
- 69. Scott-McKean J. J., Surewicz K., Choi J.-K., Ruffin V. A., Salameh A. I., Nieznanski K., Costa A. C. S., and Surewicz W. K. (2016) Soluble prion protein and its N-terminal fragment prevent impairment of synaptic plasticity by Aβ oligomers: Implications for novel therapeutic strategy in Alzheimer's disease. Neurobiol. Dis. 91, 124–131 10.1016/j.nbd.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Iraci N., Stincardini C., Barreca M. L., and Biasini E. (2015) Decoding the function of the N-terminal tail of the cellular prion protein to inspire novel therapeutic avenues for neurodegenerative diseases. Virus Res. 207, 62–68 10.1016/j.virusres.2014.10.015 [DOI] [PubMed] [Google Scholar]
- 71. Laurén J. (2014) Cellular prion protein as a therapeutic target in Alzheimer's disease. J. Alzheimer's Dis. 38, 227–244 [DOI] [PubMed] [Google Scholar]
- 72. Cohen S. I. A., Linse S., Luheshi L. M., Hellstrand E., White D. A., Rajah L., Otzen D. E., Vendruscolo M., Dobson C. M., and Knowles T. P. (2013) Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. U.S.A. 110, 9758–9763 10.1073/pnas.1218402110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Younan N. D., and Viles J. H. (2015) A comparison of three fluorophores for the detection of amyloid fibers and prefibrillar oligomeric assemblies: ThT (Thioflavin T); ANS (1-anilinonaphthalene-8-sulfonic acid); and bisANS (4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid). Biochemistry 54, 4297–4306 10.1021/acs.biochem.5b00309 [DOI] [PubMed] [Google Scholar]
- 74. Chen K. F., Peschel N., Zavodska R., Sehadova H., and Stanewsky R. (2011) QUASIMODO, a novel GPI-anchored zona pellucida protein involved in light input to the Drosophila circadian clock. Curr. Biol. 21, 719–729 10.1016/j.cub.2011.03.049,10.1016/j.sbi.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 75. Levine J. D., Funes P., Dowse H. B., and Hall J. C. (2002) Signal analysis of behavioral and molecular cycles. BMC Neurosci. 3, 1 10.1186/1471-2202-3-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Johnson E., Ringo J., Bray N., and Dowse H. (1998) Genetic and pharmacological identification of ion channels central to the Drosophila cardiac pacemaker. J. Neurogenet. 12, 1–24 10.3109/01677069809108552 [DOI] [PubMed] [Google Scholar]
- 77. Levine J. D., Funes P., Dowse H. B., and Hall J. C. (2002) Advanced analysis of a cryptochrome mutation's effects on the robustness and phase of molecular cycles in isolated peripheral tissues of Drosophila. BMC Neurosci. 3, 5 10.1186/1471-2202-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hermann C., Yoshii T., Dusik V., and Helfrich-Förster C. (2012) Neuropeptide F immunoreactive clock neurons modify evening locomotor activity and free-running period in Drosophila melanogaster. J. Comp. Neurol. 520, 970–987 10.1002/cne.22742 [DOI] [PubMed] [Google Scholar]