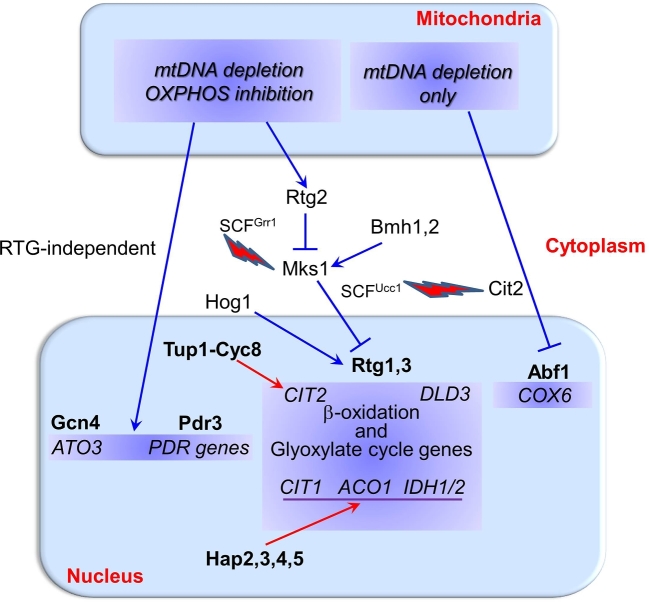
Figure 2.
Retrograde communication pathways regulating gene transcription in yeast. Key yeast genes and proteins involved in the transcriptional regulatory network of signaling pathways from mitochondria to the cytoplasm and the nucleus (see text for details). Mitochondrial dysfunction due to mtDNA depletion or OXPHOS enzyme complex inhibition can trigger retrograde pathways (blue lines), which activate-nuclear transcription to induce cell adaptation (see text for details). Dysfunctional mitochondria can activate different retrograde transcriptional responses that can be regulated by either RTG genes or alternative transcription factors and regulators, such as Abf1, Gcn4 and Pdr3. Hog1 can regulate RTG-dependent target genes. Nuclear genes highlighted in blue are upregulated by the bolded transcription factors located above them. Certain RTG-pathway target genes can also be regulated by anterograde transcription factors (see Fig. 1, red lines). The level of both retrograde target-gene products (e.g. Cit2) and regulators (e.g. Mks1) is under proteasomal degradation control, as indicated by the lightning bolts associated with the SCF ubiquitin ligase complexes.