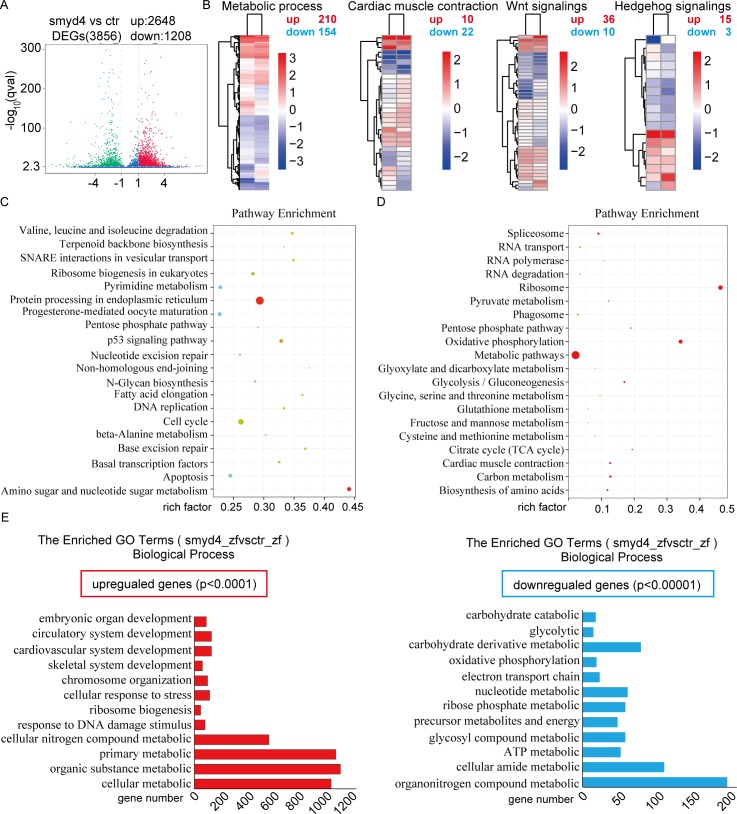
Fig 8. RNA-seq and bioinformatics analysis of MZsmyd4L544Efs*1 hearts.
RNA-seq analyses were performed with Tg(cmcl2:GFP) and MZsmyd4L544Efs*1 hearts at 72 hpf; (A) A volcano plot demonstrated that a total of 3,856 genes were differentially expressed in MZsmyd4L544Efs*1 hearts (upregulated: 2,648, downregulated: 1,208); (B) A heat-map showed that genes involved in cardiac development signaling pathways, including Wnt signaling (upregulated: 36, downregulated: 10), Hedgehog signaling (upregulated: 15, downregulated: 3), the metabolism pathway (upregulated: 210, downregulated: 154), and cardiac muscle contraction (upregulated: 10, downregulated: 22), were dysregulated in MZsmyd4L544Efs*1 hearts; (C) KEGG pathway analysis revealed a major enrichment of upregulated genes in the endoplasmic reticulum-mediated protein processing pathway; (D) KEGG pathway analysis revealed an enrichment of downregulated genes in pathways related to metabolic processes and oxidative phosphorylation; (E) Gene Ontology (GO) terms of the biological processes (BP) associated with all differentially expressed genes. The up-regulated genes function in organic substance metabolic processes, cardiovascular system development and embryonic organ development (p < 0.0001), while the downregulated genes were enriched in the carbohydrate metabolic process, the glycolytic process, and the ATP metabolic process (p < 0.00001).