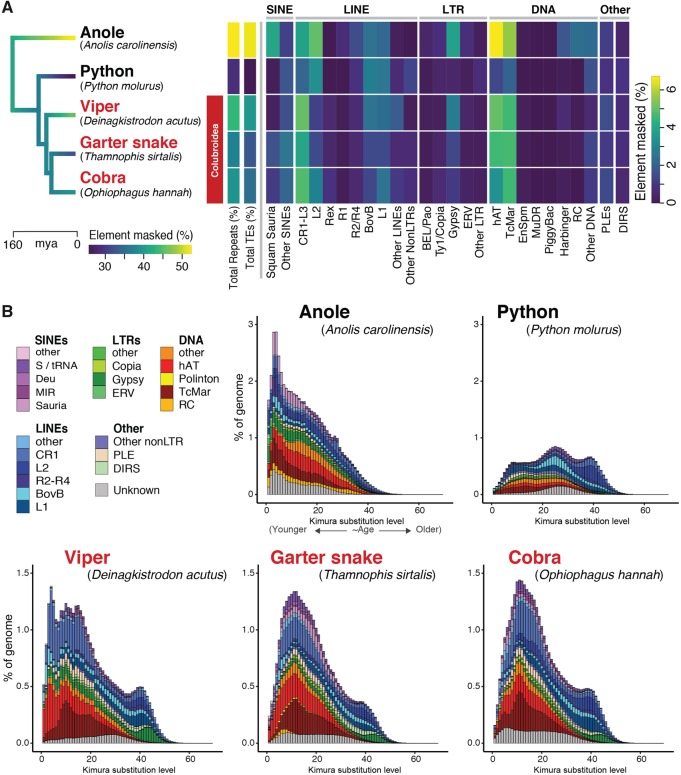
Fig. 1.
—Comparison of genomic repeat element landscapes across squamate reptiles genomes. (A) Summary of the repeat element content of the genome of Thamnophis sirtalis in comparison to other squamate reptiles. Branches on the time-calibrated consensus phylogeny are colored according to the estimated rate of genomic transposable element evolution. The associated heatmap shows the total repeat element and transposable element genomic content (%) for each taxon, as well as the genome coverage (% masked) of major components of the repeat element landscape. (B) Kimura 2-parameter distance-based TE copy divergence analysis. Genome abundance (% of genome; y-axis) is shown for each transposable element type and is clustered according to the CpG-corrected Kimura distance (K-value from 0 to 70; x-axis) from each type’s consensus sequence. Elements with low K-values are the least divergent from their respective consensus sequences, and likely represent more recent transposition events, whereas elements with higher K-values represented more degenerated copies that were likely inserted in the past.