Figure 5.
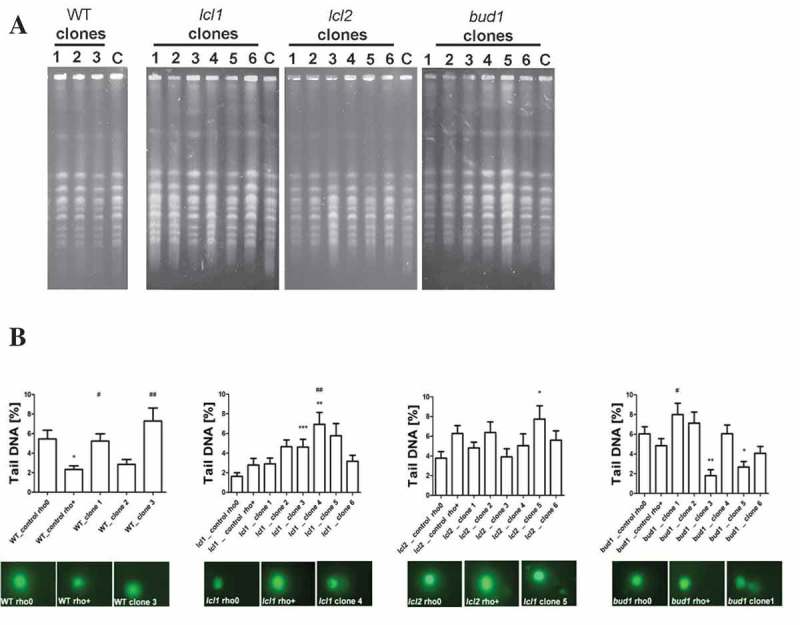
Evaluation of genomic instability at the karyotype level (a) and DNA double-strand breaks level (b) A. Electrophoretic karyotyping (PFGE separation) of haploid wild-type yeast strain BY4741, isogenic mutant strains lcl1Δ, lcl2Δ and bud1Δ (rho+ and rho°) and clones generated as daughters of an old mother. B. DNA double-strand breaks (DSBs) assessment was performed using neutral comet assay. As a DNA damage marker, the %tail DNA was used. The bars indicate SEM, n = 100, *p < 0.05, **p < 0.01, ***p < 0.001 compared to the rho+ strain, #p < 0.05, ##p < 0.01, ###p < 0.001 compared to the rho° strain (ANOVA and Dunnett’s a posteriori test). The typical micrographs are shown (bottom). DNA was visualised using YOYO-1 staining (green).
