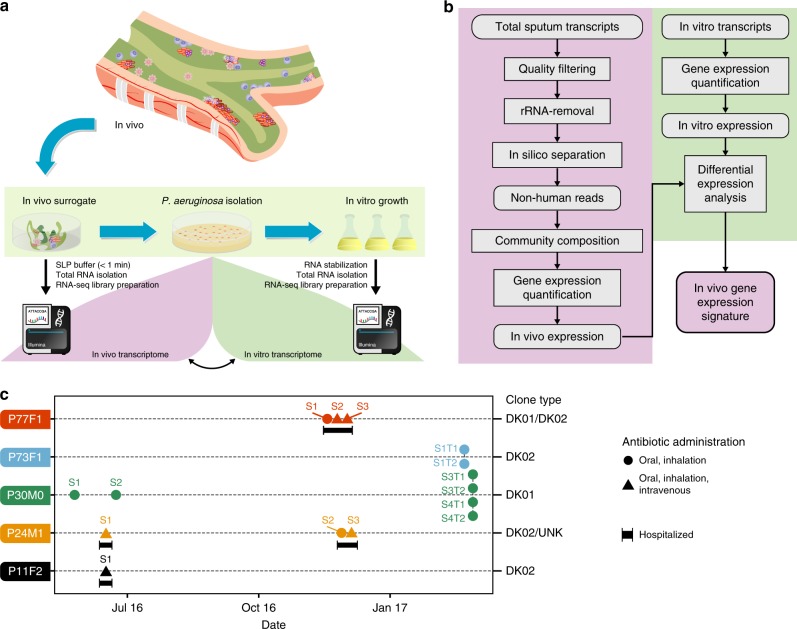
Fig. 1.
Experimental design and patients included in the study. a Schematic representation of the experimental design. In the thick mucus layer, bacterial communities thrive in a harsh environment. To capture the best representation of in vivo transcription, sputum samples were collected directly from adult CF patients followed at the Copenhagen Cystic Fibrosis Clinic at Rigshospitalet and nucleic acid content was stabilized in less than 1 min using sputum pre-lysis and preservation buffer (SLP buffer), followed by total RNA isolation, RNA-seq library preparation, and sequencing (for a complete description see Methods). From the same or a second sputum sample, we isolated, for some patients, more than ten single P. aeruginosa clones and studied their gene expression in laboratory condition (in vitro). b Simplified schematic representation of the in silico analysis workflow. Total reads were quality-filtered and any rRNA contaminant removed. Human reads were separated by mapping high-quality reads directly on human GRCh38 genome. Reads not assignable to human genome were used to evaluate microbial community composition and to assess in vivo P. aeruginosa gene expression. Transcription deriving from in vitro cultures of P. aeruginosa were used as a reference for identifying differentially gene expressed in vivo. c Overview of longitudinally collected samples. Clone types colonizing each patient are reported. Assignment of clone type was obtained by whole genome sequences from isolates obtained during this study or from previous isolates. A generic indication of antibiotic treatment and intravenous administration is provided (for a complete overview see Supplementary Table 1)