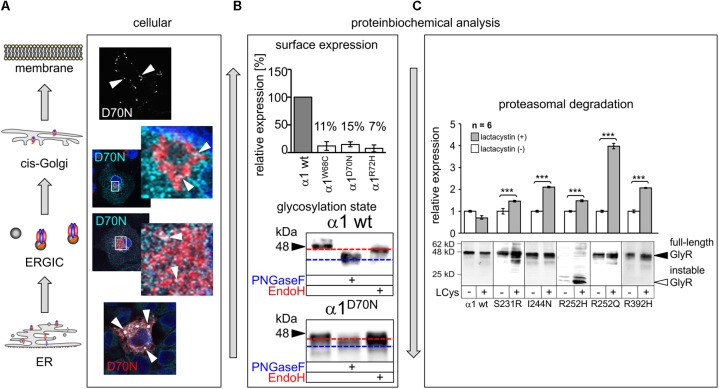
FIGURE 4.
Biogenesis and trafficking defects of GlyR α1 mutants. (A) GlyR α1 subunits carrying recessive mutations are associated with trafficking defects to the cellular surface. Compartmental analysis of transfected COS7 cells with the GlyR α1 mutant α1D70N showed very few protein dots (marked by white arrow heads) at the cellular surface (labeled before fixation with the α1 specific antibody mAb2b, 1:500). GlyR α1 protein analysis on the ER-ERGIC-cis-Golgi trafficking route determined GlyR α1D70N staining in all compartments analyzed (for ER – calnexin cyan, GlyR α1 red; ERGIC – ERGic53 red, GlyR α1 cyan; cis-Golgi – GM130 red, GlyR α1 cyan). GlyR α1D70N protein accumulation was most pronounced in the ER (large white dots). (B) GlyR protein glycosylation is a pre-requisite for ER exit. The status of protein glycosylation can be determined by digestion with glycosidases Endo H and PNGase F. PNGase F removes all N-linked oligosaccharides from glycoproteins (blue dotted line, lower images). Endo H cuts within the core of high mannose and some hybrid oligosaccharides from N-linked glycoproteins. Once a protein enters the Golgi apparatus and is further glycosylated, the protein gets resistant to Endo H digestion (red dotted line, lower images). A comparison of surface expression between GlyR wild type and mutants can be achieved by biotinylation of surface receptor protein and subsequent binding and elution from streptavidin-beads (upper image). The protein analysis is done normalized to a house keeping protein (e.g., pan-cadherin) and the assumption that the GlyR α1 wild type expression refers to 100%. (C) GlyR α1 mutants are degraded by proteasomal degradation. Using lactacystin (LCys), a proteasomal blocker, GlyR α1 mutant proteins accumulated significantly. The quantification showed significant protein increase following lactacystin treatment for all GlyR α1 mutants but not for the wild type. Images in (A–C) are were modified from Villmann et al. (2009b) and Schaefer et al. (2015). ∗∗∗p < 0.001.