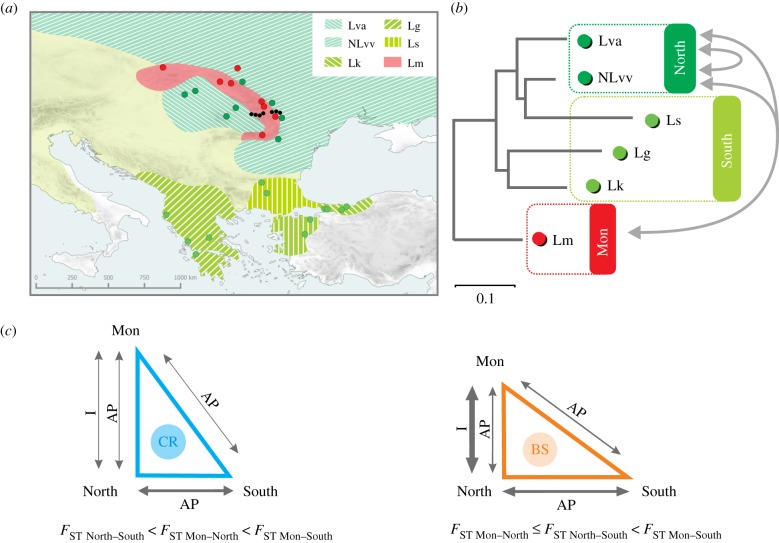
Figure 1.
Sampling and predictions. (a) Sampling sites and distribution of six lineages of the Lissotriton vulgaris species complex used in the study: L. graecus (Lg), L. kosswigi (Lk), L. montandoni (Lm), L. schmidtleri (Ls), L. v. ampelensis (Lva), northern L. v. vulgaris (NLvv); the lineages are arranged into three geographical groups: North, South and Mon; the distribution of the L. vulgaris complex lineages not included in the study is in pale green; coloured dots—localities with data for both MHC and other genes, small black dots—localities where only MHC was investigated; (b) the TreeMix drift tree depicting relationships among the six lineages [19]; Mon and North, as well as lineages within North have been connected by historical gene flow (double-headed arrows); and (c) expected differentiation between the three groups for genes under balancing selection (BS, orange) and for control genes (CR, blue) under the assumption that BS facilitates introgression; the groups are at vertices and edge lengths reflect the relative differentiation in pairwise comparisons; differentiation is determined by retention of ancestral polymorphism (AP) caused by neutral or selective processes and introgression (I); arrow thicknesses show contributions of AP and I; inequalities show the expected FST rankings. (Online version in colour.)