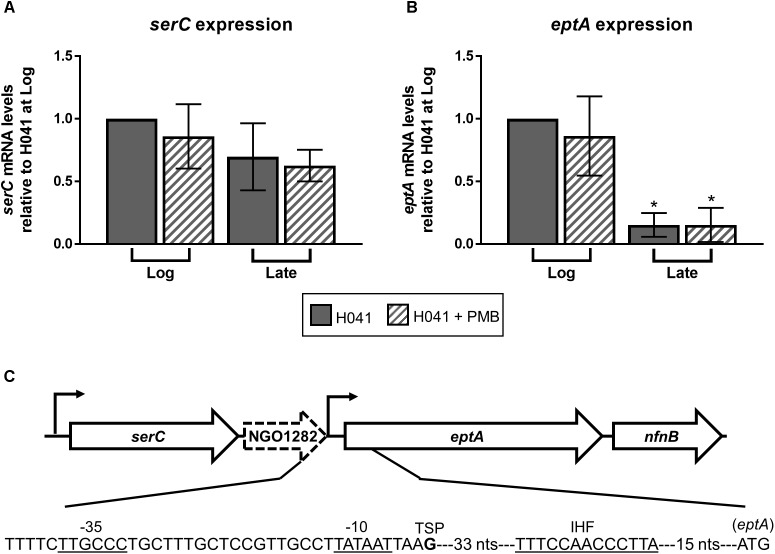
FIGURE 2.
Expression of eptA is growth phase-dependent. qRT-PCR analysis of (A) serC and (B) eptA expression in GC isolate H041 grown in GC broth with and without the addition of 1 μg/ml polymyxin B (PMB). RNA samples were collected during logarithmic growth (Log, OD600 of ∼0.4) and at the end of logarithmic growth (Late, OD600 of ∼1.0) and subject to qRT-PCR using gene-specific primers. Primers serC_qRT_F (5′-TGTTGCCTGAAGCTGTGTTG-3′) and serC_qRT_R (5′-TGTTCCGCATGATGCAGGAT-3′) were used for serC expression. Primers lptA_qRT_F (5′-GGCATCGCGATGTTGCAATA-3′) and lptA_qRT_R (5′-CACGACCGCCATATCCAATTG-3′) were used for eptA expression. 16S rRNA expression was assessed with primers 16Smai_qRTF (5′-CCATCGGTATTCCTCCACATCTCT-3′) and 16Smai_qRTR (5′-CGTAGGGTGCGAGCGTTAATC-3′). The means and standard deviations of three biological replicates are shown. The expression of serC and eptA were normalized to 16S rRNA expression at the Log timepoint without PMB treatment. Gene expression data were analyzed by ANOVA followed by Sidak’s multiple comparisons test. ∗Indicates P ≤ 0.05. (C) Organization of eptA locus and position of promoters modified from Kandler et al. (2014). The eptA -10 and -35 promoter elements, transcriptional start point (TSP) is the bolded G nucleotide, putative integration host factor binding site (IHF) based on the consensus IHF-binding site in E. coli (11 matches of 13 bp) previously described in N. gonorrhoeae (Lee et al., 2006) as indicated by the underlined sequence, and the translational start codon are shown.