Abstract
Background:
Accumulating documents have demonstrated that long noncoding RNAs (lncRNAs) play critical roles in tumorigenesis. As an lncRNA, nuclear-enriched abundant transcript 1 (NEAT1) has been identified to be involved in the progression of many types of cancers. However, the biological function of NEAT1 in cervical cancer is not fully investigated. The aim of this study was to disclose the specific biological function of lncRNA NEAT1 in cervical cancer progression.
Methods:
Quantitative real-time polymerase chain reaction (qRT-PCR) was utilized to identify the expression of lncRNA NEAT1 in the cervical cancer tissues and cell lines. All cervical cancer samples used in this study were collected from the Affiliated Suzhou Hospital of Nanjing Medical University between September 2012 and September 2017. The correlation between NEAT1 expression and the overall survival rate of cervical cancer patients was analyzed by Kaplan-Meier analysis. The effects of NEAT1 knockdown or overexpression on cell proliferation were tested by performing MTT assays and colony formation assays. Transwell assays were conducted to detect the migratory ability of cervical cancer cells, in which NEAT1 was silenced or overexpressed. Western blotting was utilized to validate whether NEAT1 promotes cervical cancer progression through activating PI3K-Akt signaling pathway.
Results:
High expression of NEAT1 predicted poor prognosis of cervical cancer patients (χ2 = 0.735, P = 0.005). Knockdown of NEAT1 decreased the number of colonies in CaSki cell from 136.667 ± 13.503 to 71.667 ± 7.506 (t = −18.76, P = 0.003) and decreased the number of colonies in HeLa cell from 128.667 ± 13.317 to 65.667 ± 7.024 (t = −5.54, P = 0.031). However, overexpression of NEAT1 increased the number of colonies in SiHa cell from 84.667 ± 12.014 to 150.667 ± 18.037 (t = 7.27, P = 0.018). Knockdown of NEAT1 decreased the migratory number of CaSki cell from 100.333 ± 9.866 to 58.333 ± 5.859 (t = −8.08, P = 0.015) and reduced the migratory number in HeLa cell from 123.667 ± 12.097 to 67.667 ± 7.095 (t = −6.03, P = 0.026). Overexpression of NEAT1 increased the migratory number of SiHa cell from 127.333 ± 16.042 to 231.333 ± 31.786 (t = 4.92, P = 0.039).
Conclusion:
NEAT1 may exert oncogenic function in cervical cancer and serve as a novel therapeutic target for cervical cancer.
Keywords: Cervical Cancer, Long Noncoding RNA Nuclear-Enriched Abundant Transcript 1, Migration, PI3K/Akt Signaling Pathway, Proliferation
摘要
背景:
越来越多的研究表明长链非编码RNA能够在肿瘤形成中发挥重要作用。长链非编码RNA NEAT1 已被证实可参与多种癌 症的发生发展。然而,NEAT1在宫颈癌中的生物学功能未有相关报道。本研究旨在揭露长链非编码RNA NEAT1在宫颈癌的癌 症发生发展过程中的生物学作用及其机制。
方法:
研究中所有组织样本均从南京医科大学附属苏州医院获取。样本收集时间为2012年9月到2017年9月。NEAT1的表达与 患者总体生存率间的关系由Kaplan-Meier生存曲线分析得出。MTT和克隆实验方法检测NEAT1干涉和过表达对细胞增殖能力 的影响。在NEAT1被干涉或过表达后,Transwell转移实验被用于检测宫颈癌细胞的转移能力。蛋白免疫印迹方法检测PI3K-Akt 信号通路相关蛋白的表达水平。转染了si-NC或pcDNA-NC的癌细胞株作为阴性对照。
结果:
NEAT1的高表达预示了宫颈癌病人的不良预后(χ2 = 7.735, P =0.005)。干涉NEAT1 将CaSki细胞的活性从 136.667 ± 13.503降低到71.667 ± 7.506(t = -18.76, P = 0.003)将HeLa细胞的活性从128.667 ± 13.317降低到65.667±7.024(t = -5.54, P = 0.031)。然而过表达NEAT1将SiHa细胞的活性84.667 ± 12.014升高到150.667 ± 18.037(t = 7.27,P = 0.018)。干涉NEAT1 后,CaSki细胞的转移数量从100.333 ± 9.866降低到58.333 ± 5.859(t = -8.08, P = 0.015),HeLa细胞的转移数量从123.667 ± 12.097 降低到67.667 ± 7.095(t = -6.03, P = 0.026)。NEAT1过表达使SiHa细胞的转移数量从127.333 ± 16.042增长到231.333 ± 31.786 (t = 4.92, P = 0.039)。
结论:
NEAT1在宫颈癌发生发展中可能发挥了癌基因的作用,NEAT1可能是宫颈癌的一个新的治疗靶点。
INTRODUCTION
Cervical cancer is the fourth most common cancer in women and the fourth prominent cause of cancer-caused mortality in women worldwide, with nearly 275,000 deaths per year.[1] Many biological factors have been demonstrated to affect the progression of cervical cancer.[2,3] Due to poor understanding of molecular mechanisms underlying the initiation and progression of cervical cancer, the prognosis of cervical cancer is still unsatisfied. At present, long noncoding RNAs (lncRNAs), with length >200 nucleotides, have been identified as critical regulators in multiple biological processes, including tumorigenesis. For example, Huang et al.[4] reported that lncRNA LINC00673 could be activated by SP1 and exerted its oncogenic properties through interacting with LSD1 and EZH2 in gastric cancer; Jin et al.[5] demonstrated that upregulation of lncRNA PlncRNA-1 promotes proliferation and induces epithelial–mesenchymal transition in prostate cancer; moreover, Li et al.[6] revealed that LINC00672 contributed p53-mediated gene suppression and promoted endometrial cancer chemosensitivity. Recently, many lncRNAs are reported to be associated with the progression and development of cervical cancer, such as TUG1, MALAT1, XIST, and ANRIL.[7,8,9,10] However, the pathophysiological processes and the specific molecular mechanisms of cervical cancer still need to be further investigated. Nuclear-enriched abundant transcript 1 (NEAT1) is a 3.2 kb novel nuclear lncRNA which is transcribed from the multiple endocrine neoplasia locus.[11] Dysregulation of NEAT1 has shown to be involved with non-small cell lung cancer, breast cancer, colorectal cancer, thyroid cancer, and pancreatic cancer.[12,13,14,15] Up to date, the biological function of NEAT1 in cervical cancer has been rarely discussed. This study focused on the biological function and mechanism of NEAT1 in cervical cancer progression.
METHODS
Ethical approval
This study had been approved by the Ethics Committee of the Affiliated Suzhou Hospital of Nanjing Medical University. Informed consent forms were obtained from all patients before this study.
Tissue samples
A total of 62 pairs of human cervical cancer tissues and adjacent normal tissues were obtained from the Affiliated Suzhou Hospital of Nanjing Medical University between September 2012 and September 2017. Patients included in the study received no chemotherapy or radiotherapy before surgery. The loss rate is about 7%. The age of all patients ranged from 38 to 60 years. The median age was 50 years. The method for sample selection was random sampling. Samples were snap-frozen and stored in liquid nitrogen as soon as they were collected.
Cell culture and transfection
Four cervical cancer cell lines (CaSki, HeLa, ME-180, and SiHa) and normal human cervical epithelial cell line (H8) were obtained from American Type Culture Collection and cultured in humidified air at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium media (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), penicillin (100 U/ml), and streptomycin (100 U/ml). Cervical cancer cell lines were transfected with 50 nmol/L Small-interfering RNA (si-RNA) specifically targeted to NEAT1 (si-NEAT1) and negative control siRNA (si-NC) using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Forty-eight-hour after transfection, total cells were harvested for total RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR) analysis.
RNA extraction and quantitative real-time polymerase chain reaction
Total RNA was isolated by TRIzol Reagent (Invitrogen) following the manufacturer's instructions. After RNA extraction, RNA samples were reversely transcribed by the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The FastStart Universal SYBR Green Master (Roche, USA) was applied for the qRT-PCR. The following primers were used to detect the expression of NEAT1 and GAPDH (internal control) – NEAT1 (sense): 5’-CTTCCTCCCTTT AACTTATCCATTC-AC-3’ and NEAT1 (antisense): 5’-CTCTTCCTCCACCATTACCAACAATAC-3’; GAPDH (sense): 5’-AGAAGGCTGGGGCTCATTTG-3’ and GAPDH (antisense): 5’-AGGGGCCATCACAGTCT TC-3’. The relative fold changes of candidate genes were analyzed using 2−ΔΔCT method.
Cell viability assay
Cell proliferation of cervical cancer cells was measured using MTT kit (Sigma, San Francisco, CA, USA). Briefly, logarithmically growing cervical cancer cells were trypsinized from culture dishes and placed at a density of 2 × 103 cells per well into a 96-well plate and transfected with indicated vector. Cell proliferation was assessed 12, 24, 48, 72, and 96 h after transfection. Spent medium was replaced with fresh medium containing MTT 0.5 mg/ml. After incubated at 37°C for another 2 h, the medium was removed and 100 μl of DMSO (Sigma) was added to each well and plates, agitating for 10 min. Absorbance was assessed at 490 nm by a spectrophotometer (BioTek, Winooski, Vermont, USA).
Colony formation assay
Cells (500 cells/well) transfected with indicated vector were plated in 6-well plates and incubated at 37°C. Two weeks later, the cells were fixed and stained with 0.1% crystal violet. The number of visible colonies was counted manually.
Cell migration assay
Cell migration was detected using Transwell chambers (8-mm pore size; Millipore, Billerica, MA, USA). In brief, 600 ml complete medium was added to the bottom chamber, transfected cells were suspended in serum-free medium, and 200 ml of the cell suspension (containing 1 × 105 cells) was placed in the upper chamber. After 48 h, the cells on the top surface of the membrane were mechanically removed using a cotton swab, and the cells on the bottom surface of the membrane were fixed in 95% ethanol and stained with 0.2% crystal violet solution. Cells adhering to the bottom surface of the membrane were counted in five randomly selected areas under microscope field. Each experiment was repeated three times.
Western blotting analysis
Cell proteins were prepared using cell lysis buffer. Equal amounts of protein (50 mg) were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes (Millipore) by electroblotting. The membranes were blocked with 5% nonfat dry milk in TBST for 1 h and then incubated with primary antibody (anti-phosphorylated PI3K [p-PI3K], total PI3K, anti-phosphorylated Akt [p-Akt], total Akt, and anti-GAPDH; Abcam, Cambridge, UK) overnight at 4°C before subsequent incubation with second antibody (Cell Signaling Technology, MA, USA) for 1 h at 37°C. Protein bands were visualized using enhanced chemiluminescence reagent (Pierce, Thermo Fisher Scientific, Inc.).
Statistical analysis
The statistical analysis was performed with SPSS software version 19.0 (IBM Corp., Armonk, NY, USA). The data were analyzed and presented as mean ± standard deviation (SD). Continuous variables were compared using the Student's t-test or the analysis of variance (ANOVA) test. Pearson χ2 test was used to evaluate the relationship between NEAT1 expression and clinical features. Survival analysis was performed using the Kaplan-Meier method, and the log-rank test was used to compare the differences between patient groups. A P < 0.05 was considered statistically significant.
RESULTS
Upregulated nuclear-enriched abundant transcript 1 expression in human cervical cancer tissues and cell lines
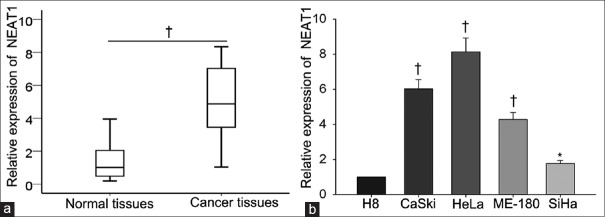
To explore whether NEAT1 was abnormal expressed in the cervical cancer tissues, qRT-PCR was performed to measure the level of NEAT1 in 62 cervical cancer tissues and corresponding normal tissues. As illustrated in Figure 1a, the expression level of NEAT1 in cervical cancer tissues was 5.155 ± 2.089 which was higher than that in normal tissues which the expression level was 1.396 ± 1.079 (t = 11.15, P = 0.007). Subsequently, the level of NEAT1 was tested in four cervical cancer cell lines (HeLa, ME-180, SiHa, and CaSki) and one normal human cervical epithelial cell line (H8) [Figure 1b]. The expression of NEAT1 in H8 cell was set as 1, the relative higher level of NEAT1 was detected in CC cells. The expression quantity of NEAT1 in CC cells was shown as follows: HeLa (8.130 ± 0.800, t = 15.43, P = 0.004), ME-180 (4.280 ± 0.400, t = 14.9, P = 0.005), SiHa (1.760 ± 0.170, t = 7.74, P = 0.016) and CaSki (6.124 ± 0.534, t =16.62, P = 0.004). The highest levels of NEAT1 were observed in CaSki and HeLa cells, while the lowest level of NEAT1 was detected in SiHa cell. Therefore, these three cells were chosen for next functional assays.
Figure 1.
Upregulated NEAT1 expression in human cervical cancer tissues and cell lines (n = 62). (a) qRT-PCR was performed to measure the level of NEAT1 in cervical cancer tissues and corresponding normal tissues. (b) Levels of NEAT1 in different cervical cancer cell lines (HeLa, ME-180, SiHa, and CaSki) and normal human cervical epithelial cell line (H8) were detected by qRT-PCR. *P < 0.05, †P < 0.01 versus control group. NEAT1: Nuclear-enriched abundant transcript 1; qRT-PCR: Quantitative real-time polymerase chain reaction.
Nuclear-enriched abundant transcript 1 correlating with the clinicopathological features and overall survival of cervical cancer patients
We assessed the correlation of NEAT1 expression with the clinicopathological parameters of 62 patients diagnosed with cervical cancer. The median value of NEAT1 in all cervical cancer tissues was used as a cutoff value, and all samples were divided into two groups (high expression group: n = 29; low expression group: n = 33). As presented in Table 1, high level of NEAT1 expression was significantly correlated with larger tumor size (χ2 = 8.058, P = 0.005), poor differentiation (χ2 = 9.226, P = 0.002), International Federation of Gynecology and Obstetrics stage (χ2 = 4.218, P = 0.040), and lymph node metastasis (χ2 = 5.156, P = 0.023) but was no significant correlation with age (χ2 = 0.827, P = 0.363) and histologic type (χ2 = 2.659, P = 0.103). Furthermore, Kaplan-Meier method analysis (log-rank test) was performed to determine the association between NEAT1 expression and overall survival of patients. As shown in Figure 2, patients with high level of NEAT1 expression had a significantly shorter overall survival than those with low level of NEAT1 (χ2 = 7.735, P = 0.005).
Table 1.
Correlation between lncRNA-NEAT1 expression and clinical features of patients with cervical cancer (n = 62)
| Variables | LncRNA-NEAT1 expression, n | χ2 | P | |
|---|---|---|---|---|
| High | Low | |||
| Age | ||||
| <50 years | 10 | 12 | 0.827 | 0.363 |
| ≥50 years | 23 | 17 | ||
| Histologic type | ||||
| SCC | 16 | 20 | 2.659 | 0.103 |
| AD/ASC | 17 | 9 | ||
| FIGO stage | ||||
| Ib–IIa | 20 | 10 | 4.218 | 0.040 |
| IIb–IIIa | 13 | 19 | ||
| Large tumor size | ||||
| <4 cm | 21 | 8 | 8.058 | 0.005 |
| ≥4 cm | 12 | 21 | ||
| Poor differentiation | ||||
| Well + moderate | 27 | 13 | 9.226 | 0.002 |
| Poor | 6 | 16 | ||
| Lymph node metastasis | ||||
| Negative | 27 | 16 | 5.156 | 0.023 |
| Positive | 6 | 13 | ||
NEAT1: Nuclear-enriched abundant transcript 1; SCC: Squamous cell carcinoma; AD: Adenocarcinoma; ASC: Adenosquamous carcinoma. FIGO: International Federation of Gynecology and Obstetrics.
Figure 2.
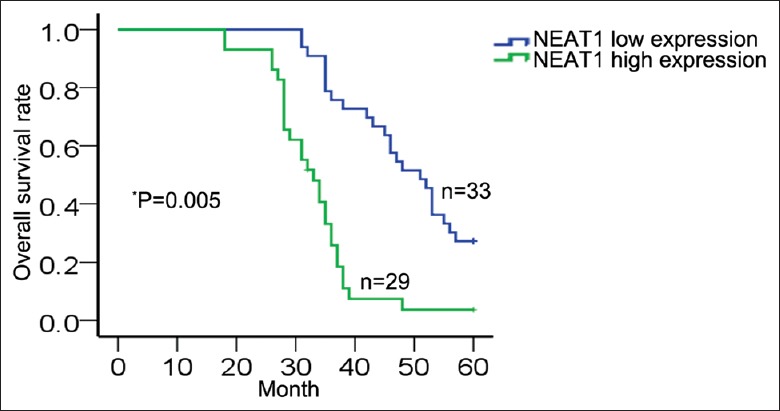
Correlation between NEAT1 and overall survival of cervical cancer patients was analyzed by Kaplan–Meier method (log-rank test: *P = 0.005, χ2 = 7.735). NEAT1: Nuclear-enriched abundant transcript 1.
Effects of nuclear-enriched abundant transcript 1 knockdown or overexpression on the proliferation and migration of cervical cancer cells
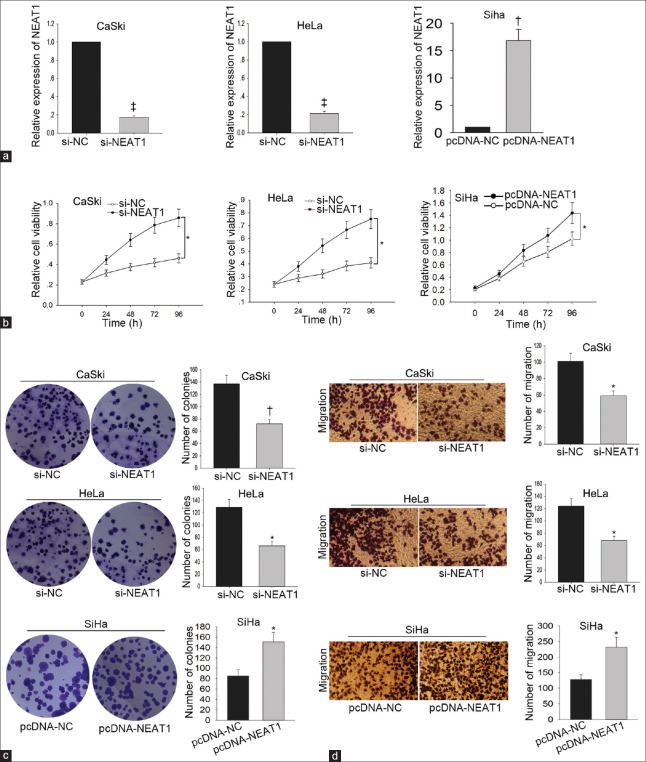
To determine the biological function of NEAT1 in cervical cancer, gain-of-function or loss-of-function experiments were performed. CaSki and HeLa cells were transfected with si-NEAT1 or si-NC. After 48 h, the transfection efficiency was obtained with qRT-PCR. The expression level of NEAT1 in cells transfected with si-NC was set as 1, and the expression level of CaSki cell was decreased to 0.174 ± 0.017 (t = −83.29, P < 0.001). Moreover, the expression level of HeLa cell was decreased to 0.213 ± 0.021 (t = −64.22, P < 0.001). NEAT1 was overexpressed in SiHa cell through transfecting with pcDNA-NEAT1 (pcDNA3.1 vector containing the whole sequence of NEAT1). The expression level of NEAT1 in SiHa cell transfected with empty vector (pcDNA-NC) was set as 1, and the expression level of NEAT1 was increased to 16.820 ± 2.050 (t = 13.38, P = 0.006) [Figure 3a]. Subsequently, functional assays were carried out in these three cells. Cell viability in CaSki cell transfected with si-NEAT1 was decreased from 0.592 ± 0.257 to 0.360 ± 0.091 (t = 3.12, P = 0.036). Cell viability in HeLa cell transfected with si-NEAT1 was decreased from 0.516 ± 0.208 to 0.328 ± 0.069 (t = 2.98, P = 0.041). Cell viability of SiHa cell transfected with pcDNA-NEAT1 was increased from 0.314 ± 0.013 to 0.728 ± 0.146 (t = 12.16, P = 0.017) [Figure 3b]. The number of colonies of CaSki cell transfected with si-NEAT1 was decreased from 136.667 ± 13.503 to 71.667 ± 7.506 (t = −18.76, P = 0.003). The number of colonies of HeLa cell transfected with si-NEAT1 was reduced from 128.667 ± 13.317 to 65.667 ± 7.024 (t = −5.54, P = 0.031). However, overexpression of NEAT1 increased the number of colonies in SiHa cell from 84.667 ± 12.014 to 150.667 ± 18.037 (t = 7.27, P = 0.018) [Figure 3c]. Knockdown of NEAT1 decreased the migratory number of CaSki cell from 100.333 ± 9.866 to 58.333 ± 5.859 (t = −8.08, P = 0.015) and of HeLa cell from 123.667 ± 12.097 to 67.667 ± 7.095 (t = −6.03, P = 0.026). Overexpression of NEAT1 increased the migratory number of SiHa cell from 127.333 ± 16.042 to 231.333 ± 31.786 (t = 4.92, P = 0.039) [Figure 3d].
Figure 3.
Effects of NEAT1 knockdown or overexpression on the proliferation and migration of cervical cancer cells. NEAT1 was silenced in CaSki and HeLa cells through transfecting with si-NEAT1. NEAT1 was overexpressed in SiHa cell through transfecting with pcDNA-NEAT1 (a); the transfection efficiency was measured by qRT-PCR. Effect of NEAT on cell proliferation ability was measured by MTT assay (b) and colony formation assay was stained with 0.1% crystal violet (c). Effect of NEAT knockdown or overexpression on cell migration capacity was measured by Transwell assay. The migratory cells were stained with 0.2% crystal violet (Original magnification ×200; d). *P < 0.05, †P < 0.01, ‡P < 0.001 versus control group. NEAT1: Nuclear-enriched abundant transcript 1; qRT-PCR: Quantitative real-time polymerase chain reaction; si-NEAT1: NEAT1-specific small-interfering RNA.
Nuclear-enriched abundant transcript 1 activating PI3K/Akt signaling pathway in cervical cancer
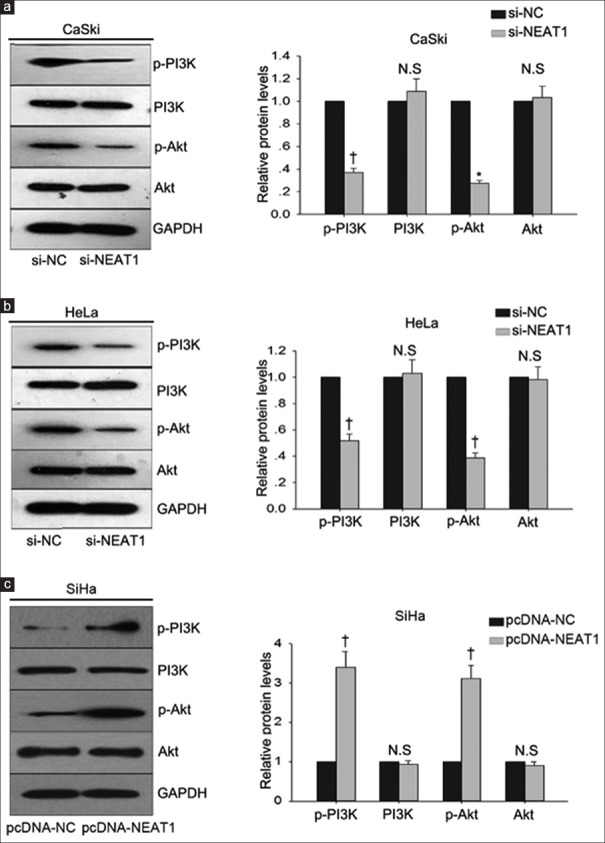
To determine the possible mechanism by which NEAT1 regulated the progression of cervical cancer cells, we analyzed the role of PI3K/Akt signal pathway in the function of NEAT1. PI3K/Akt signal pathway had been identified to be aberrantly activated in human cancers and contributed to enhanced cell proliferation and metastasis. Our findings revealed that NEAT1 positively regulated the levels of p-PI3K and p-Akt in three cervical cancer cells (CaSki, HeLa, and SiHa) [Figure 4a–4c]. The level of p-PI3K and p-Akt in CaSki or HeLa cells transfected with si-NC was defied as 1. The level of p-PI3K in CaSki cell transfected with si-NEAT1 was decreased to 0.370 ± 0.303 (t = −36.03, P = 0.001). The level of p-Akt in CaSki cell transfected with si-NEAT1 was decreased to 0.276 ± 0.301 (t = −41.67, P = 0.010). In HeLa cell transfected with si-NEAT1, the level of p-PI3K was decreased to 0.517 ± 0.515 (t = −16.25, P = 0.004), and the level of p-Akt was decreased to 0.387 ± 0.038 (t = −27.60, P = 0.001). SiHa cell was transfected with pcDNA-NEAT1 and pcDNA-NC. The level of p-PI3K and p-Akt in SiHa cell transfected with pcDNA-NC was set as 1. The level of p-PI3K in SiHa cell transfected with pcDNA-NEAT1 was increased to 3.401 ± 0.401 (t = 10.38, P = 0.009), and the level of p-Akt was increased to 3.106 ± 0.346 (t = 10.52, P = 0.009).
Figure 4.
PI3K/Akt pathway involved in the function of NEAT1. Western blotting was performed to measure the effects of NEAT1 knockdown or overexpression on the levels of p-PI3K and p-Akt in CaSki cell (a), HeLa cell (b), and SiHa cell (c). *P < 0.05, †P < 0.01 versus control group. N.S: No significance. p: Phosphorylated; NEAT1: Nuclear-enriched abundant transcript 1.
PI3K/Akt pathway involved in the function of nuclear-enriched abundant transcript 1
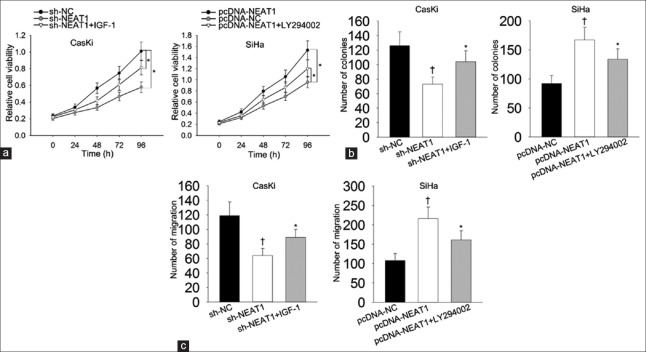
To validate whether PI3K/Akt pathway actually involved in the function of NEAT1 in cervical cancer progression, rescue assays were designed and carried out in different CC cell lines. IGF-1 and LY294002 were known as the activator and inhibitor of PI3K-Akt signaling pathway.[16,17] IGF-1 and LY294002 were separately utilized in rescue assays. Results of MTT manifested that cell proliferation inhibited by silenced NEAT1 (t = 3.05, P = 0.012) as recovered by IGF-1 (t = 4.37, P = 0.026) [Figure 5a]. However, cell proliferation promoted by NEAT1 overexpression (t = 10.33, P = 0.030) was attenuated by treating with LY294002 [Figure 5a]. Similarly, colony formation assay also revealed that cell proliferation inhibited by silenced NEAT1 was recovered by IGF-1 (F = 9.220, P = 0.015). However, cell proliferation promoted by NEAT1 overexpression was attenuated by treating with LY294002 (F = 12.775, P = 0.07) [Figure 5b]. According to the results of Transwell assay, migration ability inhibited by NEAT1 knockdown was partly recovered by treating with IGF-1 (F = 11.670, P = 0.009) [Figure 5c]. Conversely, migration ability strengthened by NEAT1 overexpression was partially weakened when cells were treated with LY294002 (F = 14.590, P = 0.050). All findings above reflected that NEAT1 positively regulates cell proliferation and migration in cervical cancer through modulating PI3K/Akt signaling pathway.
Figure 5.
PI3K/Akt pathway involved in the function of NEAT1. Effects of IGF-1 or LY294002 on si-NEAT1/pcDNA-NEAT1-mediated cell proliferation were evaluated using MTT assay (a) and colony formation assay (b). Effects of IGF-1 on si-NEAT1-mediated cell migration were measured. The effects of LY294002 on pcDNA-NEAT1-mediated cell migration were examined with Transwell assay (c). *P < 0.05, †P < 0.01 versus control group. NEAT1: Nuclear-enriched abundant transcript 1; si-NEAT1: NEAT1-specific small-interfering RNA.
DISCUSSION
Previous reports have revealed that dysregulation of NEAT1 greatly affects the prognosis of cancer patients.[18,19] This study revealed the prognostic implication of NEAT1 in cervical cancer. The high expression of NEAT1 predicted poor prognosis for cervical cancer patients. Although the clinical samples used in this study are limited, our research results still make senses. These results suggested that NEAT1 had the oncogenic property in cervical cancer. The specific biological function of NEAT1 is worth further investigating.
Many studies have revealed the oncogenic property of NEAT1 in human cancers.[20,21,22] The biological function of NEAT1 was demonstrated in the above malignant tumors by loss-of-function assays. In this study, we applied both loss-of-function and gain-of-function assays to prove the effects of NEAT1 on cellular processed in cervical cancer. Silenced NEAT1 is able to inhibited proliferation and migration of cervical cancer cells, while overexpression of NEAT1 suppressed cell proliferation and migration of cervical cancer cell. NEAT1 positively modulated cell proliferation and migration in cervical cancer. These results suggested that NEAT1 exerts as an oncogene in cervical cancer progression by promoting cell proliferation and migration. Based on the experimental results, the oncogenic role of NEAT1 was identified in cervical cancer. Although this study only studied the effects of NEAT1 on cell proliferation and migration, the experimental results still help identify the oncogenic property of NEAT1 in cervical cancer.
A lot of previous studies have demonstrated that PI3K/Akt signaling pathway involved in the lncRNAs-mediated function in many biological processes.[23,24] This study was focus on whether PI3K/Akt signaling pathway involved in NEAT1-mediated cell proliferation and migration in cervical cancer. We found that NEAT1 promoted the level of p-PI3K and p-Akt. The result indicated that NEAT1 positively affected the activation of PI3K/Akt signaling pathway. Using the activator (IGF-1) and inhibitor (LY294002) of PI3K-Akt signaling pathway, we identified that the effects of NEAT1 on cell proliferation and migration were mediated by the PI3K-Akt signaling. Taken together, we confirmed that promoted cell proliferation and migration in cervical cancer by activating PI3K/Akt signaling pathway. Therefore, NEAT1 may be considered as potential therapy target for cervical cancer. Since this study did not deeply explore the molecular mechanism by which NEAT1 regulated cervical cancer progression, we will make further investigation in the future.
Financial support and sponsorship
This study was supported by grants from the National Natural Science Foundation of China (No. 81370719 and No. 81671535), the Science Foundation of Jiangsu (No. BE2015642, and No. WSW023), the Jiangsu Key Discipline of Human Assisted Reproduction Medicine Foundation (No. FXK201749), and the Jiangsu Provincial Medical Youth Talent of the Project of Invigorating Health Care through Science, Technology, and Education (No. ZDRCA2016044).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Ning-Ning Wang
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Wang YC, Hu DY, Hu XM, Shen YQ, Meng XY, Tang H, et al. Assessing the early response of advanced cervical cancer to neoadjuvant chemotherapy using intravoxel incoherent motion diffusion-weighted magnetic resonance imaging: A pilot study. Chin Med J. 2016;129:665–71. doi: 10.4103/0366-6999.177995. doi: 10.4103/0366-6999.177995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan ZH, Zhang Y, Tian Y, Tan W, Li YH. IκB kinase b mediating the downregulation of p53 and p21 by lipopolysaccharide in human papillomavirus 16+ cervical cancer cells. Chin Med J. 2016;129:2703–7. doi: 10.4103/0366-6999.193463. doi: 10.4103/0366-6999.193463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang M, Hou J, Wang Y, Xie M, Wei C, Nie F, et al. Long noncoding RNA LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol Ther. 2017;25:1014–26. doi: 10.1016/j.ymthe.2017.01.017. doi: 10.1016/j.ymthe.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Jin Y, Cui Z, Li X, Jin X, Peng J. Upregulation of long non-coding RNA plncRNA-1 promotes proliferation and induces epithelial-mesenchymal transition in prostate cancer. Oncotarget. 2017;8:26090–9. doi: 10.18632/oncotarget.15318. doi: 10.18632/oncotarget.15318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W, Li H, Zhang L, Hu M, Li F, Deng J, et al. Long non-coding RNA LINC00672 contributes to p53 protein-mediated gene suppression and promotes endometrial cancer chemosensitivity. J Biol Chem. 2017;292:5801–13. doi: 10.1074/jbc.M116.758508. doi: 10.1074/jbc.M116.758508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y, Sun X, Mao C, Guo G, Ye S, Xu J, et al. Upregulation of long noncoding RNA TUG1 promotes cervical cancer cell proliferation and migration. Cancer Med. 2017;6:471–82. doi: 10.1002/cam4.994. doi: 10.1002/cam4.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi R, Miyagawa R, Yamashita H, Morikawa T, Okuma K, Fukayama M, et al. Increased expression of long non-coding RNA XIST predicts favorable prognosis of cervical squamous cell carcinoma subsequent to definitive chemoradiation therapy. Oncol Lett. 2016;12:3066–74. doi: 10.3892/ol.2016.5054. doi: 10.3892/ol.2016.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei R, Xue M, Zhang L, Lin Z. Long noncoding RNA MALAT1-regulated microRNA 506 modulates ovarian cancer growth by targeting iASPP. Onco Targets Ther. 2017;10:35–46. doi: 10.2147/OTT.S112686. doi: 10.2147/ott.s112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang D, Sun G, Zhang H, Tian J, Li Y. Long non-coding RNA ANRIL indicates a poor prognosis of cervical cancer and promotes carcinogenesis via PI3K/Akt pathways. Biomed Pharmacother. 2017;85:511–6. doi: 10.1016/j.biopha.2016.11.058. doi: 10.1016/j.biopha.2016.11.058. [DOI] [PubMed] [Google Scholar]
- 11.Souquere S, Beauclair G, Harper F, Fox A, Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell. 2010;21:4020–7. doi: 10.1091/mbc.E10-08-0690. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang B, Liu C, Wu Q, Zhang J, Min Q, Sheng T, et al. Long non-coding RNA NEAT1 facilitates pancreatic cancer progression through negative modulation of miR-506-3p. Biochem Biophys Res Commun. 2017;482:828–34. doi: 10.1016/j.bbrc.2016.11.120. doi: 10.1016/j.bbrc.2016.11.120. [DOI] [PubMed] [Google Scholar]
- 13.Li JH, Zhang SQ, Qiu XG, Zhang SJ, Zheng SH, Zhang DH, et al. Long non-coding RNA NEAT1 promotes malignant progression of thyroid carcinoma by regulating miRNA-214. Int J Oncol. 2017;50:708–16. doi: 10.3892/ijo.2016.3803. doi: 10.3892/ijo.2016.3803. [DOI] [PubMed] [Google Scholar]
- 14.Peng W, Wang Z, Fan H. LncRNA NEAT1 impacts cell proliferation and apoptosis of colorectal cancer via regulation of Akt signaling. Pathol Oncol Res. 2017;23:651–6. doi: 10.1007/s12253-016-0172-4. doi: 10.1007/s12253-016-0172-4. [DOI] [PubMed] [Google Scholar]
- 15.Qian K, Liu G, Tang Z, Hu Y, Fang Y, Chen Z, et al. The long non-coding RNA NEAT1 interacted with miR-101 modulates breast cancer growth by targeting EZH2. Arch Biochem Biophys. 2017;615:1–9. doi: 10.1016/j.abb.2016.12.011. doi: 10.1016/j.abb.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 16.Gan Y, He L, Yao K, Tan J, Zeng Q, Dai Y, et al. Knockdown of HMGN5 increases the chemosensitivity of human urothelial bladder cancer cells to cisplatin by targeting PI3K/Akt signaling. Oncol Lett. 2017;14:6463–70. doi: 10.3892/ol.2017.7045. doi: 10.3892/ol.2017.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin J, Sun Z, Yang F, Tang L, Chen W, Guan X, et al. MiR-19b-3p inhibits breast cancer cell proliferation and reverses saracatinib-resistance by regulating PI3K/Akt pathway. Arch Biochem Biophys. 2018;645:54–60. doi: 10.1016/j.abb.2018.03.015. doi: 10.1016/j.abb.2018.03.015. [DOI] [PubMed] [Google Scholar]
- 18.Liu X, Liang Y, Song R, Yang G, Han J, Lan Y, et al. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol Cancer. 2018;17:90. doi: 10.1186/s12943-018-0838-5. doi: 10.1186/s12943-018-0838-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Cheng C. Long noncoding RNA NEAT1 promotes the metastasis of osteosarcoma via interaction with the G9a-DNMT1-snail complex. Am J Cancer Res. 2018;8:81–90. [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Yu Y, Fan S, Luo L. Knockdown of long non-coding RNA NEAT1 inhibits proliferation and invasion and induces apoptosis of osteosarcoma by inhibiting miR-194 expression. Yonsei Med J. 2017;58:1092–100. doi: 10.3349/ymj.2017.58.6.1092. doi: 10.3349/ymj.2017.58.6.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun W, Lan X, Zhang H, Wang Z, Dong W, He L, et al. NEAT1_2 functions as a competing endogenous RNA to regulate ATAD2 expression by sponging microRNA-106b-5p in papillary thyroid cancer. Cell Death Dis. 2018;9:380. doi: 10.1038/s41419-018-0418-z. doi: 10.1038/s41419-018-0418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang X, Zhou Y, Sun AJ, Xue JL. NEAT1 contributes to breast cancer progression through modulating miR-448 and ZEB1. J Cell Physiol. 2018 doi: 10.1002/jcp.26470. [Epub ahead of print] doi: 10s.1002/jcp.26470. [DOI] [PubMed] [Google Scholar]
- 23.Lu Y, Li Y, Chai X, Kang Q, Zhao P, Xiong J, et al. Long noncoding RNA HULC promotes cell proliferation by regulating PI3K/AKT signaling pathway in chronic myeloid leukemia. Gene. 2017;607:41–6. doi: 10.1016/j.gene.2017.01.004. doi: 10.1016/j.gene.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Pan H, Jiang T, Cheng N, Wang Q, Ren S, Li X, et al. Long non-coding RNA BC087858 induces non-T790M mutation acquired resistance to EGFR-TKIs by activating PI3K/AKT and MEK/ERK pathways and EMT in non-small-cell lung cancer. Oncotarget. 2016;7:49948–60. doi: 10.18632/oncotarget.10521. doi: 10.18632/oncotarget.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]