Abstract
Rabies is a neglected viral zoonotic disease affecting humans, domestic and wild animals and is endemic in most parts of the India. Dog mediated rabies is more predominant than other forms of rabies and molecular epidemiology is poorly understood in both reservoir and susceptible hosts. In the present study, a total of 140 rabies suspected brain samples from different species of animals from different geographical regions of India were used. The samples were parallelly tested by direct fluorescent antibody test, reverse transcriptase PCR and real-time PCR. Thirty positive samples were subjected for partial nucleoprotein gene sequencing and phylogenetic analysis. On sequence and phylogenetic analysis, it was observed that all Indian rabies viruses belonged to classical rabies virus of genotype 1 of Lyssavirus and formed two distinct groups. The majority of isolates were in group-1 and are closely related to arctic/arctic like lineage, whereas group–II isolated are closely related to cosmopolitan lineage. These results indicated there is simultaneous existence of two distinct lineages of rabies viruses in Indian subcontinent. Further whole genome studies are needed for better understanding of molecular epidemiology of rabies virus circulating in animals for control and prevention of rabies in India.
Electronic supplementary material
The online version of this article (10.1007/s13337-018-0478-9) contains supplementary material, which is available to authorized users.
Keywords: Rabies, Animals, N-gene, Phylogenetic analysis, Epidemiology, India
Introduction
Rabies is a neglected zoonotic disease of mammals caused by members of the genus Lyssavirus, family Rhabdoviridae and order Mononegavirales. The causative agent, rabies virus (RABV) belongs to Phylogroup I, which also includes Australian bat lyssavirus (ABLV), Duvenhage virus (DUVV), European bat lyssaviruses 1 and 2 (EBLV1 and 2), Aravan virus (ARAV), Khujand virus (KHUV) and Irkut virus (IRKV), whereas phylogroup II includes Lagos bat virus (LBV) and Mokola virus (MOKV). The most divergent species in the genus, West Caucasian bat virus (WCBV), is not a member of either of these phylogroups (ICTV, 2017). In India, the classical RABV circulate mainly in stray dogs, which serve as the principal reservoir and transmitter of rabies to humans and other animal species [16, 17]. The RABV genome consists of five genes, which encode five proteins namely Nucleoprotein (N), Phosphoprotein (P), RNA polymerase (L), Matrix protein (M) and the Glycoprotein (G) [20]. The highly conserved N protein is critical for genome encapsidation, transcription and replication and is frequently used as a diagnostic and molecular marker for epidemiological analysis of Lyssavirus distribution regionally and globally [1, 6, 8, 12]. The genetic characterization of RABV from domestic and human origin has been studied by targeting N gene [11–15] G gene [3] and P gene [13]. However, considering the burden and endemicity of rabies in India, the molecular epidemiology of the circulating rabies virus in animals was carried out to understand the genetic makeup of rabies virus through partial N gene based sequencing and phylogenetic analysis.
Materials and methods
Ethics statement
No animals were used for this study. Suspected brain samples from different species of animals that were already dead were submitted for laboratory confirmation of rabies.
Samples
Between 2013 and 2016, a total of 140 rabies suspected brain samples from Buffalo (22), Camel (04), Cat (02), Cattle (60), Dog (48) and Horse (04) from different states of India Andhra Pradesh, Gujarat, Jammu & Kashmir, Karnataka, Kerala, Madhya Pradesh, Maharashtra, Punjab, Rajasthan, Uttar Pradesh and Uttarakhand (Table 1) were used in the study.
Table 1.
List of rabies suspected samples collected from different states in India for active surveillance
| S. no. | Name of state | No. of samples |
|---|---|---|
| 1 | Andhra Pradesh | 01 |
| 2 | Gujarat | 11 |
| 3 | Jammu & Kashmir | 01 |
| 4 | Karnataka | 58 |
| 5 | Kerala | 11 |
| 6 | Madhya Pradesh | 09 |
| 7 | Maharashtra | 14 |
| 8 | Punjab | 01 |
| 9 | Rajasthan | 09 |
| 10 | Uttar Pradesh | 23 |
| 11 | Uttarakhand | 02 |
| Total | 140 |
Screening of samples
All these samples were processed for fluorescent antibody test (FAT), reverse transcription-polymerase chain reaction (RT-PCR) and real time PCR (qPCR) in order to detect the RABV antigen/nucleic acid for providing diagnosis.
For FAT, the brain impression smears were prepared from coronal sections of brain followed by fixation in chilled acetone solution. During each run the normal brain smear (tested by all the three tests) slides were used as negative control. RABV antigens in the brain impression smears were detected by fluorescently labeled nucleoprotein gene specific monoclonal antibody (Millipore, USA) following the standard protocol of World Health Organization [9]. Briefly, the fixed brain impression smears were equilibrated to room temperature followed by washing thrice with 1X phosphate buffered saline (PBS; pH 7.2) for 5 min each with intermittent shaking. The slides were incubated with anti-rabies FITC conjugate (1:20 dilution) and incubated in humidified dark chamber at 37 °C for 1 h. The slides were washed thoroughly in PBS, mounted with aqueous mountant and examined under fluorescent microscope (Nikon, Japan) in for the presence of dusty apple green fluorescence signals.
For RT-PCR, approximately 100 mg of two coronal sections from brain were homogenized and viral RNA was extracted using QIAamp Viral RNA mini kit (QIAGEN, USA), following the manufacturer’s instructions and the extracted RNA was quantified using NanoDrop™ spectrophotometer (ND-2000, Thermoscientific, USA). Complementary DNA (cDNA) was synthesised from the viral RNA by RevertAid First Strand cDNA Synthesis (Thermo Fisher Scientific, USA), following the manufacturers protocol. A total of 1 μg RNA was used for cDNA preparation by incubating a 20 μl reaction mixture at 42 °C for 60 min followed by heat inactivation at 70 °C for 5 min. PCR was performed using N gene-specific primers (RABNF5′ TGGTCACGTGTTCAATCTCAT and RABNR 5′ATCAACAGTTTCTTCAGCCATCT3′).
Similarly for qRT-PCR was also performed using N gene specific primers [13] in 20 µl reaction volume containing 10 µl of 2 × syber green master mix of Quantifast SYBR Green (QIAGEN, Germany), 0.5 µl of 10 pM of each primers, 8.0 µl of nuclease-free water and 1 µl of cDNA. The reaction was performed for 40 cycles with 55 °C for 30 s as annealing temperature. Positive control and negative control assays were run along with NTC (no-template control).
Sequencing and phylogenetic analysis
The purified PCR products were sequenced commercially by paired-end Sanger’s di-deoxy sequencing (Xcelris Genomics, India) using gene-specific primers. The sequence data were edited and submitted to GenBank. Pairwise and multiple alignment were performed by Muscle [18]. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA7 [18]. All the published nucleotide sequences of rabies virus isolates from India and representative sequences from other parts of world which were originated from different species were included for phylogenetic analysis (Supplementary file).
Results and discussion
Detection of RABV
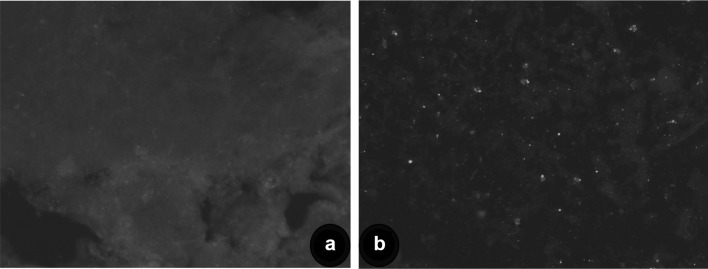
The studies on the molecular epidemiology of RABV using a substantially large number of N gene sequences of RABV from India are few. Therefore, the study was aimed to characterize Indian rabies virus from livestock and canine origin by molecular analysis in order to understand the circulating virus in livestock in India. Out of 140 rabies suspected samples, twenty-eight samples were found positive for RABV antigen. The positive brain tissues showed characteristic apple green florescent signals with varying degrees in the cytoplasm of the neurons as well as diffused in the impression smears in positive samples (Fig. 1). Conventional RT-PCR amplified the N gene sequences of rabies virus with an expected product size of 607 bp in 30 samples and all samples were also found positive by qRT-PCR without any non-specific reactions with specific melting temperature (82.24–83.11 °C). The low detection by dFAT is attributed to condition of samples received at the time of diagnosis, post-mortem changes, virus load, environmental factors etc., as reported earlier [7, 19]. The efficiency of PCR techniques for the routine diagnosis of rabies has been convincingly demonstrated, especially real-time reverse transcription PCR for RABV diagnosis [4, 5].
Fig. 1.
Detection of rabies virus by direct fluorescent antibody test (dFAT) in impression smear from brain samples showing bright apple green fluorescence in neurons (b). Negative control (a)
Sequencing and phylogenetic analysis
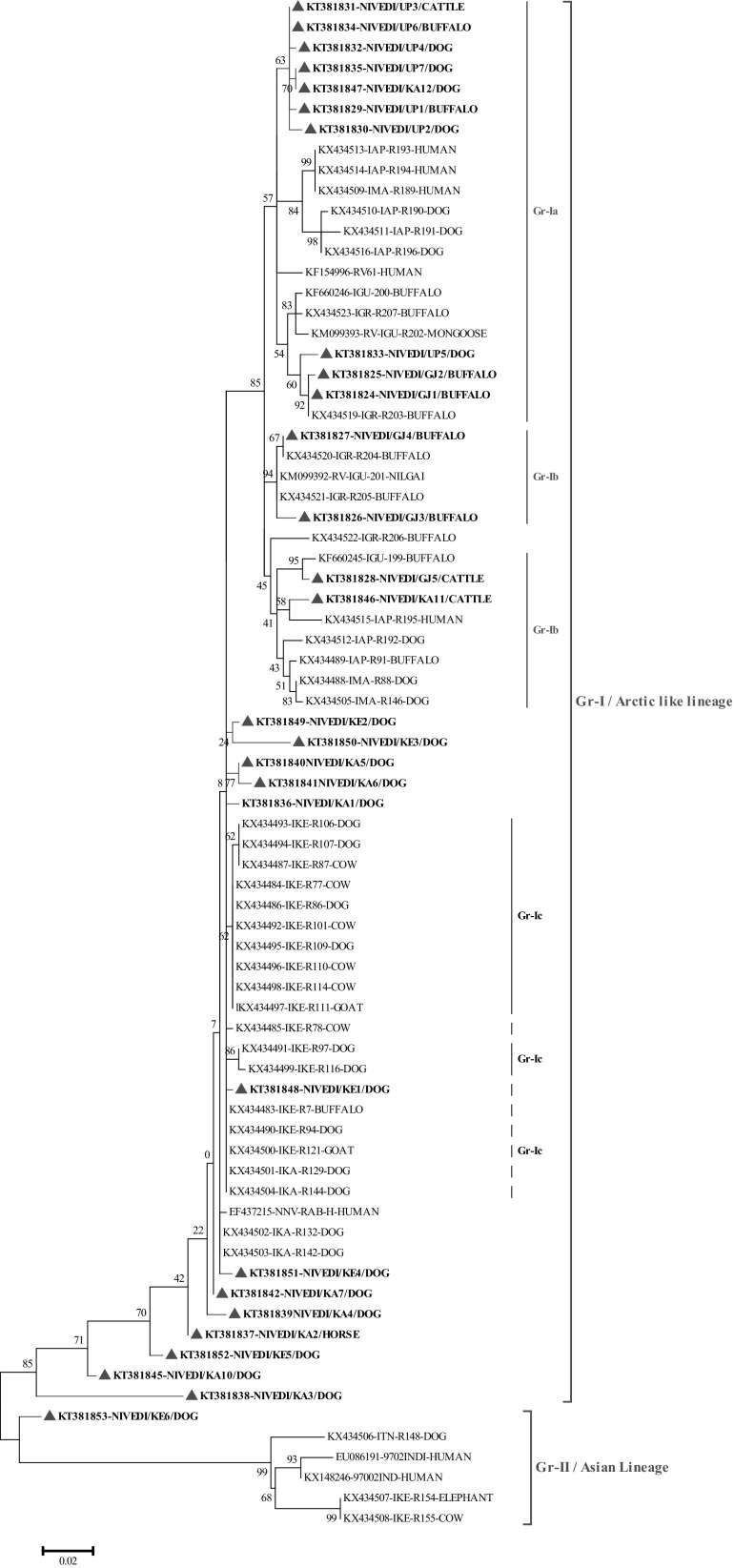
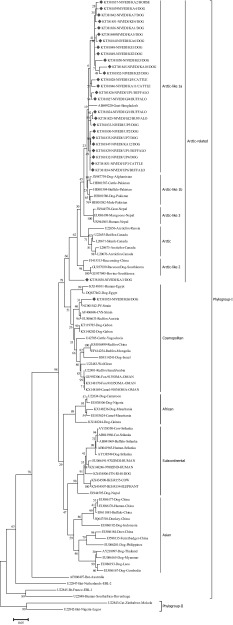
Thirty N gene nucleotide sequences of detected rabies virus in the brain samples were submitted to GenBank and obtained accession numbers (Table 2). Out of thirty partial, twenty-eight partial N gene nucleotide sequences were compared to published sequences of RABV from different species and geographical regions of India (Supplementary table). The sequences KT381843 and KT381844 were not included in phylogenetic analysis due to poor sequence coverage. The N gene-based phylogenetic tree showed two groups Group-I (Gr-I), which are closely related to arctic lineage of RABV and Group-II (Gr-II), in which there were two subgroups consisting of cosmopolitan (Gr-IIa) and subcontinental (Gr-IIb) lineage RABV. The Gr-II lineage virus (single isolate) was reported from southern part of India, where as Gr-I lineage virus are spread throughout Indian subcontinent. Further, Gr-I had three distinct subgroups namely Gr-Ia, Gr-Ib and Gr-Ic. The RABV isolates from present study typically formed different subgroups, where in all the RABV isolates from northern part of India were distributed under one single group of arctic lineage. Whereas, the RABV isolates circulating in southern part of India showed both artic and cosmopolitan lineages (Figs. 2, 3). The studies have established the presence of arctic like, cosmopolitan and subcontinental lineages among the rabies virus of India [12–14]. The majority of Indian RABV originated from Arctic like lineage (Arctic-like 1a) in India, which was confirmed by N and G gene based phylogeny [3, 10–15]. One isolate (KT381853) from the present study was grouped under the cosmopolitan lineage (Fig. 3), which supports earlier reports of only few Indian RABV identified as cosmopolitan lineage [14]. Even though, the cosmopolitan lineage has been reported from china, but in India the earlier study attributed it to laboratory cross contamination due to close relation with vaccines strain [14], but the existence of cosmopolitan lineage cannot be ruled out unless a large number of isolates are studied as the isolates in both the studies are of canine origin. Eventhough the virus were from different geographical regions of country they formed single cluster which is nearer to arctic lineage. In contrast, many reported geographically restricted distribution of RABV in India due to the physical barriers like mountain ranges and rivers which can contribute to the evolution RABVs in a region [2, 3, 11, 13].
Table 2.
Details of rabies virus isolates sequenced and used for molecular epidemiological studies in the present study
| Sl no | Isolate name | State | Species | Year | GenBank accession No. |
|---|---|---|---|---|---|
| 1 | Rab/NIVEDI/Gujarat_1 | Gujarat | Buffalo | 2014 | KT381824 |
| 2 | Rab/NIVEDI/Gujarat_2 | Gujarat | Buffalo | 2014 | KT381825 |
| 3 | Rab/NIVEDI/Gujarat_3 | Gujarat | Buffalo | 2014 | KT381826 |
| 4 | Rab/NIVEDI/Gujarat_4 | Gujarat | Buffalo | 2014 | KT381827 |
| 5 | Rab/NIVEDI/Gujarat_5 | Gujarat | Cattle | 2014 | KT381828 |
| 6 | Rab/NIVEDI/Kar_1 | Karnataka | Dog | 2013 | KT381836 |
| 7 | Rab/NIVEDI/Kar_2 | Karnataka | Equine | 2013 | KT381837 |
| 8 | Rab/NIVEDI/Kar_3 | Karnataka | Dog | 2013 | KT381838 |
| 9 | Rab/NIVEDI/Kar_4 | Karnataka | Dog | 2013 | KT381839 |
| 10 | Rab/NIVEDI/Kar_5 | Karnataka | Dog | 2013 | KT381840 |
| 11 | Rab/NIVEDI/Kar_6 | Karnataka | Dog | 2013 | KT381841 |
| 12 | Rab/NIVEDI/Kar_7 | Karnataka | Dog | 2013 | KT381842 |
| 13 | Rab/NIVEDI/Kar_8 | Karnataka | Dog | 2013 | KT381843 |
| 14 | Rab/NIVEDI/Kar_9 | Karnataka | Dog | 2013 | KT381844 |
| 15 | Rab/NIVEDI/Kar_10 | Karnataka | Dog | 2013 | KT381845 |
| 16 | Rab/NIVEDI/Kar_11 | Karnataka | Bovine | 2013 | KT381846 |
| 17 | Rab/NIVEDI/Kar_12 | Karnataka | Dog | 2013 | KT381847 |
| 18 | Rab/NIVEDI/ker_1 | Kerala | Dog | 2014 | KT381848 |
| 19 | Rab/NIVEDI/ker_2 | Kerala | Dog | 2014 | KT381849 |
| 20 | Rab/NIVEDI/ker_3 | Kerala | Dog | 2014 | KT381850 |
| 21 | Rab/NIVEDI/ker_4 | Kerala | Dog | 2014 | KT381851 |
| 22 | Rab/NIVEDI/ker_5 | Kerala | Dog | 2014 | KT381852 |
| 23 | Rab/NIVEDI/ker_6 | Kerala | Dog | 2014 | KT381853 |
| 24 | Rab/NIVEDI/UP_1 | Uttar Pradesh | Buffalo | 2014 | KT381829 |
| 25 | Rab/NIVEDI/UP_2 | Uttar Pradesh | Dog | 2014 | KT381830 |
| 26 | Rab/NIVEDI/UP_3 | Uttar Pradesh | Cattle | 2014 | KT381831 |
| 27 | Rab/NIVEDI/UP_4 | Uttar Pradesh | Dog | 2014 | KT381832 |
| 28 | Rab/NIVEDI/UP_5 | Uttar Pradesh | Dog | 2014 | KT381833 |
| 29 | Rab/NIVEDI/UP_6 | Uttar Pradesh | Buffalo | 2014 | KT381834 |
| 30 | Rab/NIVEDI/UP_7 | Uttar Pradesh | Dog | 2014 | KT381835 |
Fig. 2.
Maximum likelihood tree generated by partial nucleoprotein gene sequences showing the genetic relationship of the rabies virus isolates from India. The percentage of bootstrap values given to the left of main branch. Isolates for which the partial N gene sequence was obtained in this study are indicated by a triangle (Red: Carnivores and Green: Herbivores/Omnivores)
Fig. 3.
Maximum likelihood tree generated for nucleoprotein gene sequences showing the genetic relationship of the Indian rabies virus isolates with global RABV isolates. The percentage of bootstrap values given to the left of main branch. Isolates for which the partial N gene sequence was obtained in this study are indicated by a diamond (Red: Carnivores and Green: Herbivores/Omnivores)
Nucleotide and amino acid analysis
At nucleotide level the N gene identity of Indian RABV varied from 89.5 to 100%, while with PV strain varied from 87.2 to 96.7%. The estimated nucleotide frequencies were A = 30.05%, T = 23.45%, C = 19.91% and G = 26.59% for Indian rabies viruses. Between arctic and Asian lineages the substitution or changes were over served at 36 sites. The isolates from southern and northern India also shared high similarity ranging from 99.1 to 100% at deduced amino acid level. The arctic lineage identity of present RABV from India with neighbouring countries RABV is in accordance with previous findings that the viruses from the South Asian countries and Canada are evolved from common ancestry [10, 13, 15]. Based on N gene sequence analysis most RABV (except KT381853) similar in their genetic makeup at nucleotide and amino acid level suggesting high endemic stability of RABV in India. The deduced amino acid sequences of N gene varied at sites 14, 89 and 92 position between arctic and cosmopolitan lineages, where as an addition site at 53 position was observed between Indian and PV strain. These variations in amino acid position in combination with other genes from cosmopolitan lineage can be used to differentiate the two lineages of Indian isolates [14]. Thus, based on the present study and previous studies on domestic animal rabies featured clustering of virus isolates accroding to geographical origin irrespective of host species [11, 13, 14]. In India, dog remains as the main reservoir of rabies transmission, the possible role of wildlife in harbouring RABV needs attention. Additionally, unrestricted movement of domestic animals and no physical barrier between wild and domestic animal interface require further investigation of rabies cases especially at wild-domestic-human interface due to rise in wild animal transmitted rabies.
In conclusion, genetically similar RABV are circulating among canine reservoir species throughout India dominated arctic like lineage compared to subcontinental RABV. Hence continued mass vaccination of dogs under different rabies control programmes may lead to reduction of dog mediated rabies burden in livestock and human rabies in India.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are thankful to the Director, ICAR-NIVEDI for providing necessary facilities and SERB, DST, Government of India for funding the project (SR/FT/LS-93/2012).
References
- 1.Bourhy H, Reynes JM, Dunham EJ, Dacheux L, Larrous F, Huong VT, Xu G, Yan J, Miranda ME, Holmes EC. The origin and phylogeography of dog rabies virus. J Gen Virol. 2008;89:2673–2681. doi: 10.1099/vir.0.2008/003913-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunker K, Hampson K, Horton DL, Biek R. Integrating the landscape epidemiology and genetics of RNA viruses: rabies in domestic dogs as a model. Parasitology. 2012;139(14):1899–1913. doi: 10.1017/S003118201200090X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cherian S, Singh R, Singh KP, Manjunathareddy GB, Anjaneya Ravikumar GV, Sumithra TG, Singh RP. Phylogenetic analysis of Indian rabies virus isolates targeting the complete glycoprotein gene. Infect Genet Evol. 2015;36:333–338. doi: 10.1016/j.meegid.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 4.Dedkov VG, Deviatkin AA, Poleshchuk EM, Safonova MV, Blinova EA, Shchelkanov MY, Sidorov GN, Simonova EG, Shipulin GA. Development and evaluation of a RT-qPCR assay for fast and sensitive rabies diagnosis. Diagn Microbiol Infect Dis. 2018;90(1):18–25. doi: 10.1016/j.diagmicrobio.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Dupuis M, Brunt S, Appler K, Davis A, Rudd R. Comparison of automated quantitative reverse transcription-PCR and direct fluorescent-antibody detection for routine rabies diagnosis in the United States. J Clin Microbiol. 2015;53(9):2983–2989. doi: 10.1128/JCM.01227-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuzmin IV, Orciari LA, Arai YT, Smith JS, Hanlon CA, Kameoka Y, Rupprecht CE. Bat lyssaviruses (Aravan and Khujand) from Central Asia: phylogenetic relationships according to N, P and G gene sequences. Virus Res. 2003;97(2):65–79. doi: 10.1016/S0168-1702(03)00217-X. [DOI] [PubMed] [Google Scholar]
- 7.Manjunathareddy GB, Sumana K, Yogisharadhya R, Cherian Susan, Prajapati A, Patil SS, Balamurugan V, Singh KP, Singh R, Rahman H. Diagnosis of animal rabies: comparison of direct fluorescent antibody test (dFAT), Reverse Transcriptase-PCR and Real-time PCR. J Exp Biol Agric Sci. 2016;4(3):69–74. [Google Scholar]
- 8.Marston DA, McElhinney LM, Johnson N, Müller T, Conzelmann KK, Tordo N, Fooks AR. Comparative analysis of the full genome sequence of European bat lyssavirus type 1 and type 2 with other lyssaviruses and evidence for a conserved transcription termination and polyadenylation motif in the G-L 3′ non-translated region. J Gen Viro. 2007;88(4):1302–1314. doi: 10.1099/vir.0.82692-0. [DOI] [PubMed] [Google Scholar]
- 9.Meslin M, Koprowski H, Kaplan MM. Laboratory techniques in rabies. 4. Geneva: WHO; 1996. [Google Scholar]
- 10.Nadin-Davis SA, Turner G, Paul JP, Madhusudana SN, Wandeler AI. Emergence of Arctic-like rabies lineage in India. Emerg Infect Dis. 2007;13:111–116. doi: 10.3201/eid1301.060702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagarajan T, Mohanasubramanian B, Seshagiri EV, Nagendrakumar SB, Saseendranath MR, Satyanarayana ML, Thiagarajan D, Rangarajan PN, Srinivasan VA. Molecular epidemiology of rabies virus isolates in India. J Clin Microbiol. 2006;44:3218–3224. doi: 10.1128/JCM.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagarajan T, Nagendrakumar SB, Mohanasubramanian B, Rajalakshmi S, Hanumantha N, Ramya R, Thiagarajan D, Srinivasan VA. Phylogenetic analysis of nucleoprotein gene of dog rabies virus isolates from Southern India. Infect Genet Evol. 2009;9:976–982. doi: 10.1016/j.meegid.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Reddy GB, Singh R, Singh RP, Singh KP, Gupta PK, Desai A, Shankar SK, Ramakrishnan MA, Verma R. Molecular characterization of Indian rabies virus isolates by partial sequencing of nucleoprotein (N) and phosphoprotein (P) genes. Virus Genes. 2011;43:13–17. doi: 10.1007/s11262-011-0601-0. [DOI] [PubMed] [Google Scholar]
- 14.Reddy RV, Mohana Subramanian B, Surendra KS, Babu RP, Rana SK, Manjari KS, Srinivasan VA. Rabies virus isolates of India—simultaneous existence of two distinct evolutionary lineages. Infect Genet Evol. 2014;27:163–172. doi: 10.1016/j.meegid.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 15.Reddy RVC, Mukherjee F, Rana SK, Kanani R, Surendra KSNL, Mohana Subramanian B, Sharma GK, Srinivasan VA. Rabies virus infection in domestic buffaloes and wild animals in India. J Adv Vet Res. 2015;5(2):68–83. [Google Scholar]
- 16.Singh R, Singh KP, Cherian S, Saminathan M, Kapoor S, Manjunatha Reddy GB, Panda S, Dhama K. Rabies—epidemiology, pathogenesis, public health concerns and advances in diagnosis and control: a comprehensive review. Vet Q. 2017;37(1):212–251. doi: 10.1080/01652176.2017.1343516. [DOI] [PubMed] [Google Scholar]
- 17.Sudarshan MK, Mahendra BJ, Madhusudana SN, Rahman SA, Ashwathnarayana DH. An assessment of rabies free status of the Island of Andaman, Nicobar and Lakshadweep: results of the WHO sponsored national multicentric rabies survey. Indian J Public Health. 2006;50(1):11–14. [PubMed] [Google Scholar]
- 18.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0. Mol Biol Evol. 2015;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wacharapluesadee S, Hemachudha T. Ante and post-mortem diagnosis of rabies using nucleic acid-amplification tests. Expert Rev Mol Diagn. 2010;10:207–218. doi: 10.1586/erm.09.85. [DOI] [PubMed] [Google Scholar]
- 20.Wunner WH, Larson JK, Dietzschold B, Smith CL, Wunner WH, Larson JK, Dietzschold B, Smith CL. The molecular biology of rabies viruses. Rev Infect Dis. 1988;10(4):771–784. doi: 10.1093/clinids/10.Supplement_4.S771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.