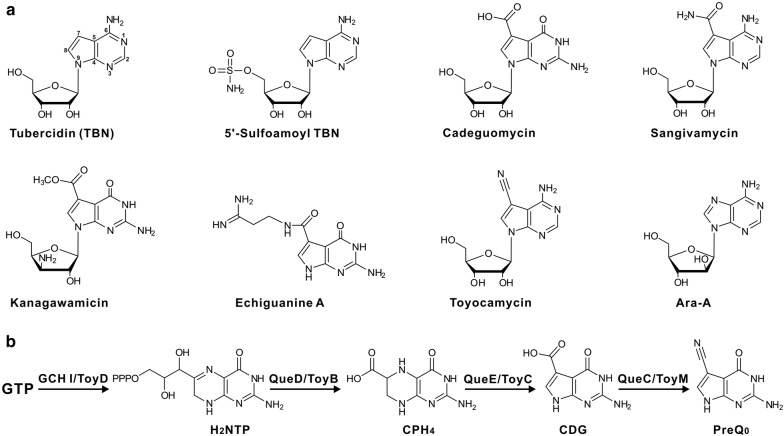
Fig. 1.
Structures of TBN and related antibiotics and the confirmed biosynthetic pathway to PreQ0. a Chemical structures of TBN and related purine nucleoside antibiotics. TBN is produced by S. tubercidicus NBRC 13090; 5′-sulfamoyl TBN is produced by S. mirabilis; cadeguomycin is produced by S. hygroscopicus; sangivamycin/toyocamycin is produced by S. rimosus ATCC 14673; kanagawamicin is produced by Actinoplanes kanagawaensis; echiguanine A is produced by Streptomyces sp. MI698-50F1; Ara-A (arabinofuranosyladenine) is produced by S. antibioticus NRRL 3238. b The confirmed biosynthetic pathway leading to PreQ0. A four-enzyme cascade for the synthesis of PreQ0 is identified in both queuosine and sangivamycin/toyocamycin biosynthetic pathways. GCH I, GTP cyclohydrolase I (ToyD); QueD/ToyB, 6-carboxy-5,6,7,8-tetrahydropterin (CPH4) synthase; H2NTP, 7, 8-dihydroneopterin-3′-triphosphate; QueE/ToyC, 7-carboxy-7-deazaguanine (CDG) synthase; QueC/ToyM, 7-cyano-7-deazaguanine (PreQ0) synthetase